initial upload
38
.gitignore
vendored
@ -1,34 +1,4 @@
|
|||||||
# ---> C++
|
.DS_Store
|
||||||
# Prerequisites
|
build/
|
||||||
*.d
|
.vscode/
|
||||||
|
tmp/
|
||||||
# Compiled Object files
|
|
||||||
*.slo
|
|
||||||
*.lo
|
|
||||||
*.o
|
|
||||||
*.obj
|
|
||||||
|
|
||||||
# Precompiled Headers
|
|
||||||
*.gch
|
|
||||||
*.pch
|
|
||||||
|
|
||||||
# Compiled Dynamic libraries
|
|
||||||
*.so
|
|
||||||
*.dylib
|
|
||||||
*.dll
|
|
||||||
|
|
||||||
# Fortran module files
|
|
||||||
*.mod
|
|
||||||
*.smod
|
|
||||||
|
|
||||||
# Compiled Static libraries
|
|
||||||
*.lai
|
|
||||||
*.la
|
|
||||||
*.a
|
|
||||||
*.lib
|
|
||||||
|
|
||||||
# Executables
|
|
||||||
*.exe
|
|
||||||
*.out
|
|
||||||
*.app
|
|
||||||
|
|
||||||
82
CMakeLists.txt
Normal file
@ -0,0 +1,82 @@
|
|||||||
|
cmake_minimum_required(VERSION 3.15.2)
|
||||||
|
# 设置项目名称与语言
|
||||||
|
project(GCTL VERSION 1.0)
|
||||||
|
# 添加配置配件编写的函数
|
||||||
|
include(CMakePackageConfigHelpers)
|
||||||
|
|
||||||
|
# 添加编译选项
|
||||||
|
option(GCTL_OPENMP "Use the OpenMP library" ON)
|
||||||
|
option(GCTL_NETCDF "Use the NetCDF library" ON)
|
||||||
|
option(GCTL_FFTW3 "Use the FFTW3 library" ON)
|
||||||
|
option(GCTL_EEMD "Use the EEMD library" ON)
|
||||||
|
option(GCTL_OPENBLAS "Use the Openblas library" OFF)
|
||||||
|
option(GCTL_CHECK_BOUNDER "Check array's index" OFF)
|
||||||
|
option(GCTL_CHECK_SIZE "Check array's size" OFF)
|
||||||
|
|
||||||
|
message(STATUS "Platform: " ${CMAKE_HOST_SYSTEM_NAME})
|
||||||
|
message(STATUS "Install prefix: " ${CMAKE_INSTALL_PREFIX})
|
||||||
|
message(STATUS "Processor: " ${CMAKE_HOST_SYSTEM_PROCESSOR})
|
||||||
|
message(STATUS "[GCTL] Use the OpenMP library: " ${GCTL_OPENMP})
|
||||||
|
message(STATUS "[GCTL] Use the NetCDF library: " ${GCTL_NETCDF})
|
||||||
|
message(STATUS "[GCTL] Use the FFTW3 library: " ${GCTL_FFTW3})
|
||||||
|
message(STATUS "[GCTL] Use the EEMD library: " ${GCTL_EEMD})
|
||||||
|
message(STATUS "[GCTL] Use the Openblas library: " ${GCTL_OPENBLAS})
|
||||||
|
message(STATUS "[GCTL] Check Bounder: " ${GCTL_CHECK_BOUNDER})
|
||||||
|
message(STATUS "[GCTL] Check Size: " ${GCTL_CHECK_SIZE})
|
||||||
|
|
||||||
|
if(GCTL_FFTW3)
|
||||||
|
if(NOT FFTW3_FOUND)
|
||||||
|
find_package(FFTW3 REQUIRED)
|
||||||
|
message(STATUS "Found FFTW3")
|
||||||
|
include_directories(${FFTW3_INC_DIR})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(GCTL_EEMD)
|
||||||
|
if(NOT EEMD_FOUND)
|
||||||
|
find_package(EEMD REQUIRED)
|
||||||
|
message(STATUS "Found EEMD")
|
||||||
|
include_directories(${EEMD_INC_DIR})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(GCTL_OPENBLAS)
|
||||||
|
if(NOT OpenBLAS_FOUND)
|
||||||
|
find_package(OpenBLAS REQUIRED)
|
||||||
|
message(STATUS "Found OpenBLAS")
|
||||||
|
include_directories(${OpenBLAS_INCLUDE_DIRS})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(GCTL_NETCDF)
|
||||||
|
if(NOT netCDF_FOUND)
|
||||||
|
find_package(netCDF REQUIRED)
|
||||||
|
include_directories(${netCDF_INCLUDE_DIR})
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(NOT netCDF_CXX_LEGACY_FOUND)
|
||||||
|
find_package(netCDF_CXX_LEGACY REQUIRED)
|
||||||
|
include_directories(${netCDF_CXX_LEGACY_INC_DIR})
|
||||||
|
endif()
|
||||||
|
|
||||||
|
message(STATUS "Found NetCDF")
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(GCTL_OPENMP)
|
||||||
|
if(NOT OpenMP_CXX_FOUND)
|
||||||
|
find_package(OpenMP REQUIRED)
|
||||||
|
set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} ${OpenMP_CXX_FLAGS}")
|
||||||
|
set(CMAKE_EXE_LINKER_FLAGS "${CMAKE_EXE_LINKER_FLAGS} ${OpenMP_EXE_LINKER_FLAGS}")
|
||||||
|
include_directories(${OpenMP_CXX_INCLUDE_DIRS})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
# 加入一个头文件配置,让cmake对源码进行操作
|
||||||
|
configure_file(
|
||||||
|
"${PROJECT_SOURCE_DIR}/config.h.in"
|
||||||
|
"${PROJECT_SOURCE_DIR}/lib/gctl_config.h"
|
||||||
|
)
|
||||||
|
|
||||||
|
# 添加库源文件地址
|
||||||
|
add_subdirectory(lib)
|
||||||
|
add_subdirectory(example)
|
||||||
76
GCTLConfig.cmake.in
Normal file
@ -0,0 +1,76 @@
|
|||||||
|
@PACKAGE_INIT@
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_VERSION "@PROJECT_VERSION@")
|
||||||
|
set_and_check(@PROJECT_NAME@_INSTALL_PREFIX "${PACKAGE_PREFIX_DIR}")
|
||||||
|
set_and_check(@PROJECT_NAME@_INC_DIR "${PACKAGE_PREFIX_DIR}/include")
|
||||||
|
set_and_check(@PROJECT_NAME@_INCLUDE_DIR "${PACKAGE_PREFIX_DIR}/include")
|
||||||
|
set_and_check(@PROJECT_NAME@_LIB_DIR "${PACKAGE_PREFIX_DIR}/lib")
|
||||||
|
set_and_check(@PROJECT_NAME@_LIBRARY_DIR "${PACKAGE_PREFIX_DIR}/lib")
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_LIB gctl)
|
||||||
|
set(@PROJECT_NAME@_LIBRARY gctl)
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_NETCDF @GCTL_NETCDF@)
|
||||||
|
set(@PROJECT_NAME@_FFTW3 @GCTL_FFTW3@)
|
||||||
|
set(@PROJECT_NAME@_EEMD @GCTL_EEMD@)
|
||||||
|
set(@PROJECT_NAME@_OPENMP @GCTL_OPENMP@)
|
||||||
|
set(@PROJECT_NAME@_OPENBLAS @GCTL_OPENBLAS@)
|
||||||
|
set(@PROJECT_NAME@_CHECK_BOUNDER @GCTL_CHECK_BOUNDER@)
|
||||||
|
set(@PROJECT_NAME@_CHECK_SIZE @GCTL_CHECK_SIZE@)
|
||||||
|
|
||||||
|
message(STATUS "[GCTL] Use the NetCDF library: " @GCTL_NETCDF@)
|
||||||
|
message(STATUS "[GCTL] Use the FFTW3 library: " @GCTL_FFTW3@)
|
||||||
|
message(STATUS "[GCTL] Use the EEMD library: " @GCTL_EEMD@)
|
||||||
|
message(STATUS "[GCTL] Use the OpenMP library: " @GCTL_OPENMP@)
|
||||||
|
message(STATUS "[GCTL] Use the Openblas library: " @GCTL_OPENBLAS@)
|
||||||
|
message(STATUS "[GCTL] Check Bounder: " @GCTL_CHECK_BOUNDER@)
|
||||||
|
message(STATUS "[GCTL] Check Size: " @GCTL_CHECK_SIZE@)
|
||||||
|
|
||||||
|
set(CMAKE_CXX_STANDARD 11)
|
||||||
|
|
||||||
|
if(@PROJECT_NAME@_NETCDF)
|
||||||
|
if(NOT netCDF_FOUND)
|
||||||
|
find_package(netCDF REQUIRED)
|
||||||
|
include_directories(${netCDF_INCLUDE_DIR})
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(NOT netCDF_CXX_LEGACY_FOUND)
|
||||||
|
find_package(netCDF_CXX_LEGACY REQUIRED)
|
||||||
|
include_directories(${netCDF_CXX_LEGACY_INC_DIR})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(@PROJECT_NAME@_FFTW3)
|
||||||
|
if(NOT FFTW3_FOUND)
|
||||||
|
find_package(FFTW3 REQUIRED)
|
||||||
|
include_directories(${FFTW3_INC_DIR})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(@PROJECT_NAME@_EEMD)
|
||||||
|
if(NOT EEMD_FOUND)
|
||||||
|
find_package(EEMD REQUIRED)
|
||||||
|
include_directories(${EEMD_INC_DIR})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(@PROJECT_NAME@_OPENBLAS)
|
||||||
|
if(NOT OpenBLAS_FOUND)
|
||||||
|
find_package(OpenBLAS REQUIRED)
|
||||||
|
include_directories(${OpenBLAS_INCLUDE_DIRS})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
if(@PROJECT_NAME@_OPENMP)
|
||||||
|
if(NOT OpenMP_CXX_FOUND)
|
||||||
|
find_package(OpenMP REQUIRED)
|
||||||
|
set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} ${OpenMP_CXX_FLAGS}")
|
||||||
|
set(CMAKE_EXE_LINKER_FLAGS "${CMAKE_EXE_LINKER_FLAGS} ${OpenMP_EXE_LINKER_FLAGS}")
|
||||||
|
include_directories(${OpenMP_CXX_INCLUDE_DIRS})
|
||||||
|
endif()
|
||||||
|
endif()
|
||||||
|
|
||||||
|
# include target information
|
||||||
|
include("${CMAKE_CURRENT_LIST_DIR}/@PROJECT_NAME@Targets.cmake")
|
||||||
|
|
||||||
|
check_required_components(@PROJECT_NAME@)
|
||||||
522
LICENSE
Normal file
@ -0,0 +1,522 @@
|
|||||||
|
GCTL License
|
||||||
|
--------------
|
||||||
|
|
||||||
|
GCTL is distributed under a dual licensing scheme. You can redistribute
|
||||||
|
it and/or modify it under the terms of the GNU Lesser General Public
|
||||||
|
License as published by the Free Software Foundation, either version 2
|
||||||
|
of the License, or (at your option) any later version. A copy of the GNU
|
||||||
|
Lesser General Public License is reproduced below.
|
||||||
|
|
||||||
|
If the terms and conditions of the LGPL v.2. would prevent you from using
|
||||||
|
the GCTL, please consider the option to obtain a commercial license for a
|
||||||
|
fee. These licenses are offered by the GCTL's original author. As a rule,
|
||||||
|
licenses are provided "as-is", unlimited in time for a one time fee. Please
|
||||||
|
send corresponding requests to: yizhang-geo@zju.edu.cn. Please do not forget
|
||||||
|
to include some description of your company and the realm of its activities.
|
||||||
|
Also add information on how to contact you by electronic and paper mail.
|
||||||
|
|
||||||
|
=====================================================================
|
||||||
|
GNU LESSER GENERAL PUBLIC LICENSE
|
||||||
|
Version 2.1, February 1999
|
||||||
|
|
||||||
|
Copyright (C) 1991, 1999 Free Software Foundation, Inc.
|
||||||
|
51 Franklin Street, Fifth Floor, Boston, MA 02110-1301 USA
|
||||||
|
Everyone is permitted to copy and distribute verbatim copies
|
||||||
|
of this license document, but changing it is not allowed.
|
||||||
|
|
||||||
|
[This is the first released version of the Lesser GPL. It also counts
|
||||||
|
as the successor of the GNU Library Public License, version 2, hence
|
||||||
|
the version number 2.1.]
|
||||||
|
|
||||||
|
Preamble
|
||||||
|
|
||||||
|
The licenses for most software are designed to take away your
|
||||||
|
freedom to share and change it. By contrast, the GNU General Public
|
||||||
|
Licenses are intended to guarantee your freedom to share and change
|
||||||
|
free software--to make sure the software is free for all its users.
|
||||||
|
|
||||||
|
This license, the Lesser General Public License, applies to some
|
||||||
|
specially designated software packages--typically libraries--of the
|
||||||
|
Free Software Foundation and other authors who decide to use it. You
|
||||||
|
can use it too, but we suggest you first think carefully about whether
|
||||||
|
this license or the ordinary General Public License is the better
|
||||||
|
strategy to use in any particular case, based on the explanations below.
|
||||||
|
|
||||||
|
When we speak of free software, we are referring to freedom of use,
|
||||||
|
not price. Our General Public Licenses are designed to make sure that
|
||||||
|
you have the freedom to distribute copies of free software (and charge
|
||||||
|
for this service if you wish); that you receive source code or can get
|
||||||
|
it if you want it; that you can change the software and use pieces of
|
||||||
|
it in new free programs; and that you are informed that you can do
|
||||||
|
these things.
|
||||||
|
|
||||||
|
To protect your rights, we need to make restrictions that forbid
|
||||||
|
distributors to deny you these rights or to ask you to surrender these
|
||||||
|
rights. These restrictions translate to certain responsibilities for
|
||||||
|
you if you distribute copies of the library or if you modify it.
|
||||||
|
|
||||||
|
For example, if you distribute copies of the library, whether gratis
|
||||||
|
or for a fee, you must give the recipients all the rights that we gave
|
||||||
|
you. You must make sure that they, too, receive or can get the source
|
||||||
|
code. If you link other code with the library, you must provide
|
||||||
|
complete object files to the recipients, so that they can relink them
|
||||||
|
with the library after making changes to the library and recompiling
|
||||||
|
it. And you must show them these terms so they know their rights.
|
||||||
|
|
||||||
|
We protect your rights with a two-step method: (1) we copyright the
|
||||||
|
library, and (2) we offer you this license, which gives you legal
|
||||||
|
permission to copy, distribute and/or modify the library.
|
||||||
|
|
||||||
|
To protect each distributor, we want to make it very clear that
|
||||||
|
there is no warranty for the free library. Also, if the library is
|
||||||
|
modified by someone else and passed on, the recipients should know
|
||||||
|
that what they have is not the original version, so that the original
|
||||||
|
author's reputation will not be affected by problems that might be
|
||||||
|
introduced by others.
|
||||||
|
|
||||||
|
Finally, software patents pose a constant threat to the existence of
|
||||||
|
any free program. We wish to make sure that a company cannot
|
||||||
|
effectively restrict the users of a free program by obtaining a
|
||||||
|
restrictive license from a patent holder. Therefore, we insist that
|
||||||
|
any patent license obtained for a version of the library must be
|
||||||
|
consistent with the full freedom of use specified in this license.
|
||||||
|
|
||||||
|
Most GNU software, including some libraries, is covered by the
|
||||||
|
ordinary GNU General Public License. This license, the GNU Lesser
|
||||||
|
General Public License, applies to certain designated libraries, and
|
||||||
|
is quite different from the ordinary General Public License. We use
|
||||||
|
this license for certain libraries in order to permit linking those
|
||||||
|
libraries into non-free programs.
|
||||||
|
|
||||||
|
When a program is linked with a library, whether statically or using
|
||||||
|
a shared library, the combination of the two is legally speaking a
|
||||||
|
combined work, a derivative of the original library. The ordinary
|
||||||
|
General Public License therefore permits such linking only if the
|
||||||
|
entire combination fits its criteria of freedom. The Lesser General
|
||||||
|
Public License permits more lax criteria for linking other code with
|
||||||
|
the library.
|
||||||
|
|
||||||
|
We call this license the "Lesser" General Public License because it
|
||||||
|
does Less to protect the user's freedom than the ordinary General
|
||||||
|
Public License. It also provides other free software developers Less
|
||||||
|
of an advantage over competing non-free programs. These disadvantages
|
||||||
|
are the reason we use the ordinary General Public License for many
|
||||||
|
libraries. However, the Lesser license provides advantages in certain
|
||||||
|
special circumstances.
|
||||||
|
|
||||||
|
For example, on rare occasions, there may be a special need to
|
||||||
|
encourage the widest possible use of a certain library, so that it becomes
|
||||||
|
a de-facto standard. To achieve this, non-free programs must be
|
||||||
|
allowed to use the library. A more frequent case is that a free
|
||||||
|
library does the same job as widely used non-free libraries. In this
|
||||||
|
case, there is little to gain by limiting the free library to free
|
||||||
|
software only, so we use the Lesser General Public License.
|
||||||
|
|
||||||
|
In other cases, permission to use a particular library in non-free
|
||||||
|
programs enables a greater number of people to use a large body of
|
||||||
|
free software. For example, permission to use the GNU C Library in
|
||||||
|
non-free programs enables many more people to use the whole GNU
|
||||||
|
operating system, as well as its variant, the GNU/Linux operating
|
||||||
|
system.
|
||||||
|
|
||||||
|
Although the Lesser General Public License is Less protective of the
|
||||||
|
users' freedom, it does ensure that the user of a program that is
|
||||||
|
linked with the Library has the freedom and the wherewithal to run
|
||||||
|
that program using a modified version of the Library.
|
||||||
|
|
||||||
|
The precise terms and conditions for copying, distribution and
|
||||||
|
modification follow. Pay close attention to the difference between a
|
||||||
|
"work based on the library" and a "work that uses the library". The
|
||||||
|
former contains code derived from the library, whereas the latter must
|
||||||
|
be combined with the library in order to run.
|
||||||
|
|
||||||
|
GNU LESSER GENERAL PUBLIC LICENSE
|
||||||
|
TERMS AND CONDITIONS FOR COPYING, DISTRIBUTION AND MODIFICATION
|
||||||
|
|
||||||
|
0. This License Agreement applies to any software library or other
|
||||||
|
program which contains a notice placed by the copyright holder or
|
||||||
|
other authorized party saying it may be distributed under the terms of
|
||||||
|
this Lesser General Public License (also called "this License").
|
||||||
|
Each licensee is addressed as "you".
|
||||||
|
|
||||||
|
A "library" means a collection of software functions and/or data
|
||||||
|
prepared so as to be conveniently linked with application programs
|
||||||
|
(which use some of those functions and data) to form executables.
|
||||||
|
|
||||||
|
The "Library", below, refers to any such software library or work
|
||||||
|
which has been distributed under these terms. A "work based on the
|
||||||
|
Library" means either the Library or any derivative work under
|
||||||
|
copyright law: that is to say, a work containing the Library or a
|
||||||
|
portion of it, either verbatim or with modifications and/or translated
|
||||||
|
straightforwardly into another language. (Hereinafter, translation is
|
||||||
|
included without limitation in the term "modification".)
|
||||||
|
|
||||||
|
"Source code" for a work means the preferred form of the work for
|
||||||
|
making modifications to it. For a library, complete source code means
|
||||||
|
all the source code for all modules it contains, plus any associated
|
||||||
|
interface definition files, plus the scripts used to control compilation
|
||||||
|
and installation of the library.
|
||||||
|
|
||||||
|
Activities other than copying, distribution and modification are not
|
||||||
|
covered by this License; they are outside its scope. The act of
|
||||||
|
running a program using the Library is not restricted, and output from
|
||||||
|
such a program is covered only if its contents constitute a work based
|
||||||
|
on the Library (independent of the use of the Library in a tool for
|
||||||
|
writing it). Whether that is true depends on what the Library does
|
||||||
|
and what the program that uses the Library does.
|
||||||
|
|
||||||
|
1. You may copy and distribute verbatim copies of the Library's
|
||||||
|
complete source code as you receive it, in any medium, provided that
|
||||||
|
you conspicuously and appropriately publish on each copy an
|
||||||
|
appropriate copyright notice and disclaimer of warranty; keep intact
|
||||||
|
all the notices that refer to this License and to the absence of any
|
||||||
|
warranty; and distribute a copy of this License along with the
|
||||||
|
Library.
|
||||||
|
|
||||||
|
You may charge a fee for the physical act of transferring a copy,
|
||||||
|
and you may at your option offer warranty protection in exchange for a
|
||||||
|
fee.
|
||||||
|
|
||||||
|
2. You may modify your copy or copies of the Library or any portion
|
||||||
|
of it, thus forming a work based on the Library, and copy and
|
||||||
|
distribute such modifications or work under the terms of Section 1
|
||||||
|
above, provided that you also meet all of these conditions:
|
||||||
|
|
||||||
|
a) The modified work must itself be a software library.
|
||||||
|
|
||||||
|
b) You must cause the files modified to carry prominent notices
|
||||||
|
stating that you changed the files and the date of any change.
|
||||||
|
|
||||||
|
c) You must cause the whole of the work to be licensed at no
|
||||||
|
charge to all third parties under the terms of this License.
|
||||||
|
|
||||||
|
d) If a facility in the modified Library refers to a function or a
|
||||||
|
table of data to be supplied by an application program that uses
|
||||||
|
the facility, other than as an argument passed when the facility
|
||||||
|
is invoked, then you must make a good faith effort to ensure that,
|
||||||
|
in the event an application does not supply such function or
|
||||||
|
table, the facility still operates, and performs whatever part of
|
||||||
|
its purpose remains meaningful.
|
||||||
|
|
||||||
|
(For example, a function in a library to compute square roots has
|
||||||
|
a purpose that is entirely well-defined independent of the
|
||||||
|
application. Therefore, Subsection 2d requires that any
|
||||||
|
application-supplied function or table used by this function must
|
||||||
|
be optional: if the application does not supply it, the square
|
||||||
|
root function must still compute square roots.)
|
||||||
|
|
||||||
|
These requirements apply to the modified work as a whole. If
|
||||||
|
identifiable sections of that work are not derived from the Library,
|
||||||
|
and can be reasonably considered independent and separate works in
|
||||||
|
themselves, then this License, and its terms, do not apply to those
|
||||||
|
sections when you distribute them as separate works. But when you
|
||||||
|
distribute the same sections as part of a whole which is a work based
|
||||||
|
on the Library, the distribution of the whole must be on the terms of
|
||||||
|
this License, whose permissions for other licensees extend to the
|
||||||
|
entire whole, and thus to each and every part regardless of who wrote
|
||||||
|
it.
|
||||||
|
|
||||||
|
Thus, it is not the intent of this section to claim rights or contest
|
||||||
|
your rights to work written entirely by you; rather, the intent is to
|
||||||
|
exercise the right to control the distribution of derivative or
|
||||||
|
collective works based on the Library.
|
||||||
|
|
||||||
|
In addition, mere aggregation of another work not based on the Library
|
||||||
|
with the Library (or with a work based on the Library) on a volume of
|
||||||
|
a storage or distribution medium does not bring the other work under
|
||||||
|
the scope of this License.
|
||||||
|
|
||||||
|
3. You may opt to apply the terms of the ordinary GNU General Public
|
||||||
|
License instead of this License to a given copy of the Library. To do
|
||||||
|
this, you must alter all the notices that refer to this License, so
|
||||||
|
that they refer to the ordinary GNU General Public License, version 2,
|
||||||
|
instead of to this License. (If a newer version than version 2 of the
|
||||||
|
ordinary GNU General Public License has appeared, then you can specify
|
||||||
|
that version instead if you wish.) Do not make any other change in
|
||||||
|
these notices.
|
||||||
|
|
||||||
|
Once this change is made in a given copy, it is irreversible for
|
||||||
|
that copy, so the ordinary GNU General Public License applies to all
|
||||||
|
subsequent copies and derivative works made from that copy.
|
||||||
|
|
||||||
|
This option is useful when you wish to copy part of the code of
|
||||||
|
the Library into a program that is not a library.
|
||||||
|
|
||||||
|
4. You may copy and distribute the Library (or a portion or
|
||||||
|
derivative of it, under Section 2) in object code or executable form
|
||||||
|
under the terms of Sections 1 and 2 above provided that you accompany
|
||||||
|
it with the complete corresponding machine-readable source code, which
|
||||||
|
must be distributed under the terms of Sections 1 and 2 above on a
|
||||||
|
medium customarily used for software interchange.
|
||||||
|
|
||||||
|
If distribution of object code is made by offering access to copy
|
||||||
|
from a designated place, then offering equivalent access to copy the
|
||||||
|
source code from the same place satisfies the requirement to
|
||||||
|
distribute the source code, even though third parties are not
|
||||||
|
compelled to copy the source along with the object code.
|
||||||
|
|
||||||
|
5. A program that contains no derivative of any portion of the
|
||||||
|
Library, but is designed to work with the Library by being compiled or
|
||||||
|
linked with it, is called a "work that uses the Library". Such a
|
||||||
|
work, in isolation, is not a derivative work of the Library, and
|
||||||
|
therefore falls outside the scope of this License.
|
||||||
|
|
||||||
|
However, linking a "work that uses the Library" with the Library
|
||||||
|
creates an executable that is a derivative of the Library (because it
|
||||||
|
contains portions of the Library), rather than a "work that uses the
|
||||||
|
library". The executable is therefore covered by this License.
|
||||||
|
Section 6 states terms for distribution of such executables.
|
||||||
|
|
||||||
|
When a "work that uses the Library" uses material from a header file
|
||||||
|
that is part of the Library, the object code for the work may be a
|
||||||
|
derivative work of the Library even though the source code is not.
|
||||||
|
Whether this is true is especially significant if the work can be
|
||||||
|
linked without the Library, or if the work is itself a library. The
|
||||||
|
threshold for this to be true is not precisely defined by law.
|
||||||
|
|
||||||
|
If such an object file uses only numerical parameters, data
|
||||||
|
structure layouts and accessors, and small macros and small inline
|
||||||
|
functions (ten lines or less in length), then the use of the object
|
||||||
|
file is unrestricted, regardless of whether it is legally a derivative
|
||||||
|
work. (Executables containing this object code plus portions of the
|
||||||
|
Library will still fall under Section 6.)
|
||||||
|
|
||||||
|
Otherwise, if the work is a derivative of the Library, you may
|
||||||
|
distribute the object code for the work under the terms of Section 6.
|
||||||
|
Any executables containing that work also fall under Section 6,
|
||||||
|
whether or not they are linked directly with the Library itself.
|
||||||
|
|
||||||
|
6. As an exception to the Sections above, you may also combine or
|
||||||
|
link a "work that uses the Library" with the Library to produce a
|
||||||
|
work containing portions of the Library, and distribute that work
|
||||||
|
under terms of your choice, provided that the terms permit
|
||||||
|
modification of the work for the customer's own use and reverse
|
||||||
|
engineering for debugging such modifications.
|
||||||
|
|
||||||
|
You must give prominent notice with each copy of the work that the
|
||||||
|
Library is used in it and that the Library and its use are covered by
|
||||||
|
this License. You must supply a copy of this License. If the work
|
||||||
|
during execution displays copyright notices, you must include the
|
||||||
|
copyright notice for the Library among them, as well as a reference
|
||||||
|
directing the user to the copy of this License. Also, you must do one
|
||||||
|
of these things:
|
||||||
|
|
||||||
|
a) Accompany the work with the complete corresponding
|
||||||
|
machine-readable source code for the Library including whatever
|
||||||
|
changes were used in the work (which must be distributed under
|
||||||
|
Sections 1 and 2 above); and, if the work is an executable linked
|
||||||
|
with the Library, with the complete machine-readable "work that
|
||||||
|
uses the Library", as object code and/or source code, so that the
|
||||||
|
user can modify the Library and then relink to produce a modified
|
||||||
|
executable containing the modified Library. (It is understood
|
||||||
|
that the user who changes the contents of definitions files in the
|
||||||
|
Library will not necessarily be able to recompile the application
|
||||||
|
to use the modified definitions.)
|
||||||
|
|
||||||
|
b) Use a suitable shared library mechanism for linking with the
|
||||||
|
Library. A suitable mechanism is one that (1) uses at run time a
|
||||||
|
copy of the library already present on the user's computer system,
|
||||||
|
rather than copying library functions into the executable, and (2)
|
||||||
|
will operate properly with a modified version of the library, if
|
||||||
|
the user installs one, as long as the modified version is
|
||||||
|
interface-compatible with the version that the work was made with.
|
||||||
|
|
||||||
|
c) Accompany the work with a written offer, valid for at
|
||||||
|
least three years, to give the same user the materials
|
||||||
|
specified in Subsection 6a, above, for a charge no more
|
||||||
|
than the cost of performing this distribution.
|
||||||
|
|
||||||
|
d) If distribution of the work is made by offering access to copy
|
||||||
|
from a designated place, offer equivalent access to copy the above
|
||||||
|
specified materials from the same place.
|
||||||
|
|
||||||
|
e) Verify that the user has already received a copy of these
|
||||||
|
materials or that you have already sent this user a copy.
|
||||||
|
|
||||||
|
For an executable, the required form of the "work that uses the
|
||||||
|
Library" must include any data and utility programs needed for
|
||||||
|
reproducing the executable from it. However, as a special exception,
|
||||||
|
the materials to be distributed need not include anything that is
|
||||||
|
normally distributed (in either source or binary form) with the major
|
||||||
|
components (compiler, kernel, and so on) of the operating system on
|
||||||
|
which the executable runs, unless that component itself accompanies
|
||||||
|
the executable.
|
||||||
|
|
||||||
|
It may happen that this requirement contradicts the license
|
||||||
|
restrictions of other proprietary libraries that do not normally
|
||||||
|
accompany the operating system. Such a contradiction means you cannot
|
||||||
|
use both them and the Library together in an executable that you
|
||||||
|
distribute.
|
||||||
|
|
||||||
|
7. You may place library facilities that are a work based on the
|
||||||
|
Library side-by-side in a single library together with other library
|
||||||
|
facilities not covered by this License, and distribute such a combined
|
||||||
|
library, provided that the separate distribution of the work based on
|
||||||
|
the Library and of the other library facilities is otherwise
|
||||||
|
permitted, and provided that you do these two things:
|
||||||
|
|
||||||
|
a) Accompany the combined library with a copy of the same work
|
||||||
|
based on the Library, uncombined with any other library
|
||||||
|
facilities. This must be distributed under the terms of the
|
||||||
|
Sections above.
|
||||||
|
|
||||||
|
b) Give prominent notice with the combined library of the fact
|
||||||
|
that part of it is a work based on the Library, and explaining
|
||||||
|
where to find the accompanying uncombined form of the same work.
|
||||||
|
|
||||||
|
8. You may not copy, modify, sublicense, link with, or distribute
|
||||||
|
the Library except as expressly provided under this License. Any
|
||||||
|
attempt otherwise to copy, modify, sublicense, link with, or
|
||||||
|
distribute the Library is void, and will automatically terminate your
|
||||||
|
rights under this License. However, parties who have received copies,
|
||||||
|
or rights, from you under this License will not have their licenses
|
||||||
|
terminated so long as such parties remain in full compliance.
|
||||||
|
|
||||||
|
9. You are not required to accept this License, since you have not
|
||||||
|
signed it. However, nothing else grants you permission to modify or
|
||||||
|
distribute the Library or its derivative works. These actions are
|
||||||
|
prohibited by law if you do not accept this License. Therefore, by
|
||||||
|
modifying or distributing the Library (or any work based on the
|
||||||
|
Library), you indicate your acceptance of this License to do so, and
|
||||||
|
all its terms and conditions for copying, distributing or modifying
|
||||||
|
the Library or works based on it.
|
||||||
|
|
||||||
|
10. Each time you redistribute the Library (or any work based on the
|
||||||
|
Library), the recipient automatically receives a license from the
|
||||||
|
original licensor to copy, distribute, link with or modify the Library
|
||||||
|
subject to these terms and conditions. You may not impose any further
|
||||||
|
restrictions on the recipients' exercise of the rights granted herein.
|
||||||
|
You are not responsible for enforcing compliance by third parties with
|
||||||
|
this License.
|
||||||
|
|
||||||
|
11. If, as a consequence of a court judgment or allegation of patent
|
||||||
|
infringement or for any other reason (not limited to patent issues),
|
||||||
|
conditions are imposed on you (whether by court order, agreement or
|
||||||
|
otherwise) that contradict the conditions of this License, they do not
|
||||||
|
excuse you from the conditions of this License. If you cannot
|
||||||
|
distribute so as to satisfy simultaneously your obligations under this
|
||||||
|
License and any other pertinent obligations, then as a consequence you
|
||||||
|
may not distribute the Library at all. For example, if a patent
|
||||||
|
license would not permit royalty-free redistribution of the Library by
|
||||||
|
all those who receive copies directly or indirectly through you, then
|
||||||
|
the only way you could satisfy both it and this License would be to
|
||||||
|
refrain entirely from distribution of the Library.
|
||||||
|
|
||||||
|
If any portion of this section is held invalid or unenforceable under any
|
||||||
|
particular circumstance, the balance of the section is intended to apply,
|
||||||
|
and the section as a whole is intended to apply in other circumstances.
|
||||||
|
|
||||||
|
It is not the purpose of this section to induce you to infringe any
|
||||||
|
patents or other property right claims or to contest validity of any
|
||||||
|
such claims; this section has the sole purpose of protecting the
|
||||||
|
integrity of the free software distribution system which is
|
||||||
|
implemented by public license practices. Many people have made
|
||||||
|
generous contributions to the wide range of software distributed
|
||||||
|
through that system in reliance on consistent application of that
|
||||||
|
system; it is up to the author/donor to decide if he or she is willing
|
||||||
|
to distribute software through any other system and a licensee cannot
|
||||||
|
impose that choice.
|
||||||
|
|
||||||
|
This section is intended to make thoroughly clear what is believed to
|
||||||
|
be a consequence of the rest of this License.
|
||||||
|
|
||||||
|
12. If the distribution and/or use of the Library is restricted in
|
||||||
|
certain countries either by patents or by copyrighted interfaces, the
|
||||||
|
original copyright holder who places the Library under this License may add
|
||||||
|
an explicit geographical distribution limitation excluding those countries,
|
||||||
|
so that distribution is permitted only in or among countries not thus
|
||||||
|
excluded. In such case, this License incorporates the limitation as if
|
||||||
|
written in the body of this License.
|
||||||
|
|
||||||
|
13. The Free Software Foundation may publish revised and/or new
|
||||||
|
versions of the Lesser General Public License from time to time.
|
||||||
|
Such new versions will be similar in spirit to the present version,
|
||||||
|
but may differ in detail to address new problems or concerns.
|
||||||
|
|
||||||
|
Each version is given a distinguishing version number. If the Library
|
||||||
|
specifies a version number of this License which applies to it and
|
||||||
|
"any later version", you have the option of following the terms and
|
||||||
|
conditions either of that version or of any later version published by
|
||||||
|
the Free Software Foundation. If the Library does not specify a
|
||||||
|
license version number, you may choose any version ever published by
|
||||||
|
the Free Software Foundation.
|
||||||
|
|
||||||
|
14. If you wish to incorporate parts of the Library into other free
|
||||||
|
programs whose distribution conditions are incompatible with these,
|
||||||
|
write to the author to ask for permission. For software which is
|
||||||
|
copyrighted by the Free Software Foundation, write to the Free
|
||||||
|
Software Foundation; we sometimes make exceptions for this. Our
|
||||||
|
decision will be guided by the two goals of preserving the free status
|
||||||
|
of all derivatives of our free software and of promoting the sharing
|
||||||
|
and reuse of software generally.
|
||||||
|
|
||||||
|
NO WARRANTY
|
||||||
|
|
||||||
|
15. BECAUSE THE LIBRARY IS LICENSED FREE OF CHARGE, THERE IS NO
|
||||||
|
WARRANTY FOR THE LIBRARY, TO THE EXTENT PERMITTED BY APPLICABLE LAW.
|
||||||
|
EXCEPT WHEN OTHERWISE STATED IN WRITING THE COPYRIGHT HOLDERS AND/OR
|
||||||
|
OTHER PARTIES PROVIDE THE LIBRARY "AS IS" WITHOUT WARRANTY OF ANY
|
||||||
|
KIND, EITHER EXPRESSED OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, THE
|
||||||
|
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR
|
||||||
|
PURPOSE. THE ENTIRE RISK AS TO THE QUALITY AND PERFORMANCE OF THE
|
||||||
|
LIBRARY IS WITH YOU. SHOULD THE LIBRARY PROVE DEFECTIVE, YOU ASSUME
|
||||||
|
THE COST OF ALL NECESSARY SERVICING, REPAIR OR CORRECTION.
|
||||||
|
|
||||||
|
16. IN NO EVENT UNLESS REQUIRED BY APPLICABLE LAW OR AGREED TO IN
|
||||||
|
WRITING WILL ANY COPYRIGHT HOLDER, OR ANY OTHER PARTY WHO MAY MODIFY
|
||||||
|
AND/OR REDISTRIBUTE THE LIBRARY AS PERMITTED ABOVE, BE LIABLE TO YOU
|
||||||
|
FOR DAMAGES, INCLUDING ANY GENERAL, SPECIAL, INCIDENTAL OR
|
||||||
|
CONSEQUENTIAL DAMAGES ARISING OUT OF THE USE OR INABILITY TO USE THE
|
||||||
|
LIBRARY (INCLUDING BUT NOT LIMITED TO LOSS OF DATA OR DATA BEING
|
||||||
|
RENDERED INACCURATE OR LOSSES SUSTAINED BY YOU OR THIRD PARTIES OR A
|
||||||
|
FAILURE OF THE LIBRARY TO OPERATE WITH ANY OTHER SOFTWARE), EVEN IF
|
||||||
|
SUCH HOLDER OR OTHER PARTY HAS BEEN ADVISED OF THE POSSIBILITY OF SUCH
|
||||||
|
DAMAGES.
|
||||||
|
|
||||||
|
END OF TERMS AND CONDITIONS
|
||||||
|
|
||||||
|
How to Apply These Terms to Your New Libraries
|
||||||
|
|
||||||
|
If you develop a new library, and you want it to be of the greatest
|
||||||
|
possible use to the public, we recommend making it free software that
|
||||||
|
everyone can redistribute and change. You can do so by permitting
|
||||||
|
redistribution under these terms (or, alternatively, under the terms of the
|
||||||
|
ordinary General Public License).
|
||||||
|
|
||||||
|
To apply these terms, attach the following notices to the library. It is
|
||||||
|
safest to attach them to the start of each source file to most effectively
|
||||||
|
convey the exclusion of warranty; and each file should have at least the
|
||||||
|
"copyright" line and a pointer to where the full notice is found.
|
||||||
|
|
||||||
|
<one line to give the library's name and a brief idea of what it does.>
|
||||||
|
Copyright (C) <year> <name of author>
|
||||||
|
|
||||||
|
This library is free software; you can redistribute it and/or
|
||||||
|
modify it under the terms of the GNU Lesser General Public
|
||||||
|
License as published by the Free Software Foundation; either
|
||||||
|
version 2.1 of the License, or (at your option) any later version.
|
||||||
|
|
||||||
|
This library is distributed in the hope that it will be useful,
|
||||||
|
but WITHOUT ANY WARRANTY; without even the implied warranty of
|
||||||
|
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU
|
||||||
|
Lesser General Public License for more details.
|
||||||
|
|
||||||
|
You should have received a copy of the GNU Lesser General Public
|
||||||
|
License along with this library; if not, write to the Free Software
|
||||||
|
Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA 02110-1301
|
||||||
|
USA
|
||||||
|
|
||||||
|
Also add information on how to contact you by electronic and paper mail.
|
||||||
|
|
||||||
|
You should also get your employer (if you work as a programmer) or your
|
||||||
|
school, if any, to sign a "copyright disclaimer" for the library, if
|
||||||
|
necessary. Here is a sample; alter the names:
|
||||||
|
|
||||||
|
Yoyodyne, Inc., hereby disclaims all copyright interest in the
|
||||||
|
library `Frob' (a library for tweaking knobs) written by James Random
|
||||||
|
Hacker.
|
||||||
|
|
||||||
|
<signature of Ty Coon>, 1 April 1990
|
||||||
|
Ty Coon, President of Vice
|
||||||
|
|
||||||
|
That's all there is to it!
|
||||||
42
README.md
@ -1,2 +1,42 @@
|
|||||||
# gctl
|
# Geophysical Computational Tools & Library (GCTL)
|
||||||
|
|
||||||
|
GCTL is a package of computational tools and C++ library for geophysical studies. The complete package is composed of a core library and additional libraries and command line tools. A full list of GCTL's libraries are listed as:
|
||||||
|
|
||||||
|
1. **gctl**: the core library (stored at this repository);
|
||||||
|
2. **gctl_potential**: library of the algorithms for working with the potential field data (i.e., gravitational and magnetic data);
|
||||||
|
3. **gctl_seismic**: library of the algorithms for working with the seismic/earthquake data;
|
||||||
|
4. **gctl_elecmag**: library of the algorithms for working with the elecmagnetic/magnetotelluric data;
|
||||||
|
5. **gctl_optimization**: library of the optimization algorithms;
|
||||||
|
6. **gctl_ai**: library of the artificial intelligence algorithms;
|
||||||
|
6. **gctl_graphic**: library for data visualization;
|
||||||
|
7. **gctl_examples**: executable examples;
|
||||||
|
8. **gctl_toolkits**: command line tools.
|
||||||
|
|
||||||
|
## Dependences
|
||||||
|
|
||||||
|
There are several third-party libraries that could be employed by the GCTL during the compilation to enable extral functionalities. The inclusion of the exterior libraries are controlled using the CMake compile options as `-D<option>=ON|OFF`. And the availiable options are:
|
||||||
|
|
||||||
|
1. **GCTL_OPENMP**: Enable OpenMP support. The default is ON;
|
||||||
|
2. **GCTL_NETCDF**: Use the netCDF libraries for reading and writing .nc files. This option requires the netCDF and netCDF_CXX libraries. The default is ON;
|
||||||
|
3. **GCTL_WAVELIB**: Use the WaveLIB for preforming the wavelet processes. The default is ON;
|
||||||
|
4. **GCTL_FFTW3**: Use the FFTW library for preforming the Fourier transformations. The default is ON;
|
||||||
|
5. **GCTL_OPENBLAS**: Use the openblas library for linear algebras. The default is OFF;
|
||||||
|
6. **GCTL_EIGEN**: Use the Eigen3 library. This is not a compiling option but could be used in applications to enable Eigen3 support.
|
||||||
|
7. **GCTL\_CHECK_BOUNDER**: Check indexing validities in all array-like operations. This may affect the computational efficiency. The default is OFF;
|
||||||
|
8. **GCTL\_CHECK_SIZE**: Check sizes in all array-like operations. This may affect the computational efficiency. The default is OFF.
|
||||||
|
|
||||||
|
## Installation
|
||||||
|
|
||||||
|
You can use the enclosed shell script 'installer' to install.
|
||||||
|
|
||||||
|
```
|
||||||
|
./installer configure && ./installer build && ./installer install
|
||||||
|
```
|
||||||
|
|
||||||
|
For more information of the 'installer', use:
|
||||||
|
|
||||||
|
```
|
||||||
|
./installer help
|
||||||
|
```
|
||||||
|
|
||||||
|
|
||||||
|
|||||||
7
config.h.in
Normal file
@ -0,0 +1,7 @@
|
|||||||
|
#cmakedefine GCTL_OPENMP
|
||||||
|
#cmakedefine GCTL_NETCDF
|
||||||
|
#cmakedefine GCTL_FFTW3
|
||||||
|
#cmakedefine GCTL_EEMD
|
||||||
|
#cmakedefine GCTL_OPENBLAS
|
||||||
|
#cmakedefine GCTL_CHECK_BOUNDER
|
||||||
|
#cmakedefine GCTL_CHECK_SIZE
|
||||||
19
dep/magnetic_tesseroids/CMakeLists.txt
Normal file
@ -0,0 +1,19 @@
|
|||||||
|
cmake_minimum_required(VERSION 3.15.2)
|
||||||
|
# 设置项目名称与语言
|
||||||
|
project(LibMagTess VERSION 1.0)
|
||||||
|
# 添加配置配件编写的函数
|
||||||
|
include(CMakePackageConfigHelpers)
|
||||||
|
|
||||||
|
if(${CMAKE_HOST_SYSTEM_NAME} STREQUAL "Windows")
|
||||||
|
set(CMAKE_INSTALL_PREFIX D:/Library)
|
||||||
|
else()
|
||||||
|
set(CMAKE_INSTALL_PREFIX /opt/stow/magtess)
|
||||||
|
endif()
|
||||||
|
|
||||||
|
message(STATUS "Platform: " ${CMAKE_HOST_SYSTEM_NAME})
|
||||||
|
message(STATUS "Install prefix: " ${CMAKE_INSTALL_PREFIX})
|
||||||
|
message(STATUS "Build type: " ${CMAKE_BUILD_TYPE})
|
||||||
|
|
||||||
|
# 添加库源文件地址
|
||||||
|
add_subdirectory(lib)
|
||||||
|
add_subdirectory(toolkits)
|
||||||
33
dep/magnetic_tesseroids/LICENSE.txt
Normal file
@ -0,0 +1,33 @@
|
|||||||
|
BSD 2-Clause License
|
||||||
|
|
||||||
|
Copyright (c) 2012-2015, Leonardo Uieda
|
||||||
|
Copyright (c) 2017-2020, Eldar Baykiev
|
||||||
|
All rights reserved.
|
||||||
|
|
||||||
|
Redistribution and use in source and binary forms, with or without
|
||||||
|
modification, are permitted provided that the following conditions are met:
|
||||||
|
|
||||||
|
1. Redistributions of source code must retain the above copyright notice, this
|
||||||
|
list of conditions and the following disclaimer.
|
||||||
|
|
||||||
|
2. Redistributions in binary form must reproduce the above copyright notice,
|
||||||
|
this list of conditions and the following disclaimer in the documentation
|
||||||
|
and/or other materials provided with the distribution.
|
||||||
|
|
||||||
|
3. Neither the name of Leonardo Uieda nor the names of any contributors*
|
||||||
|
may be used to endorse or promote products derived from this software
|
||||||
|
without specific prior written permission.
|
||||||
|
|
||||||
|
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
|
||||||
|
AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
|
||||||
|
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
|
||||||
|
DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
|
||||||
|
FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
|
||||||
|
DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
|
||||||
|
SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
|
||||||
|
CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
|
||||||
|
OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
|
||||||
|
OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
|
||||||
|
|
||||||
|
* Contributors to the original tesseroids source and binary forms, which are
|
||||||
|
under BSD 3-Clause License
|
||||||
15
dep/magnetic_tesseroids/LibMagTessConfig.cmake.in
Normal file
@ -0,0 +1,15 @@
|
|||||||
|
@PACKAGE_INIT@
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_Version "@PROJECT_VERSION@")
|
||||||
|
set_and_check(@PROJECT_NAME@_INSTALL_PREFIX "${PACKAGE_PREFIX_DIR}")
|
||||||
|
set_and_check(@PROJECT_NAME@_INC_DIR "${PACKAGE_PREFIX_DIR}/include")
|
||||||
|
set_and_check(@PROJECT_NAME@_INCULDE_DIR "${PACKAGE_PREFIX_DIR}/include")
|
||||||
|
set_and_check(@PROJECT_NAME@_LIB_DIR "${PACKAGE_PREFIX_DIR}/lib")
|
||||||
|
set_and_check(@PROJECT_NAME@_LIBRARY_DIR "${PACKAGE_PREFIX_DIR}/lib")
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_LIB magtess)
|
||||||
|
set(@PROJECT_NAME@_LIBRARY magtess)
|
||||||
|
set(@PROJECT_NAME@_FOUND 1)
|
||||||
|
|
||||||
|
# include target information
|
||||||
|
include("${CMAKE_CURRENT_LIST_DIR}/@PROJECT_NAME@Targets.cmake")
|
||||||
122
dep/magnetic_tesseroids/README.md
Normal file
@ -0,0 +1,122 @@
|
|||||||
|
# Forward modeling of magnetic field in spherical coordinates
|
||||||
|
|
||||||
|
Magnetic tesseroids is a collection of command-line tools for modelling of the magnetic field with spherical prisms (tesseroids) used as magnetic sources.
|
||||||
|
|
||||||
|
To cite _magnetic tesseroids_ in publications, please use our paper published in Computers & Geosciences:
|
||||||
|
|
||||||
|
>**Eldar Baykiev**, **Jörg Ebbing**, **Marco Brönner**, **Karl Fabian**, Forward modeling magnetic fields of induced and remanent magnetization in the lithosphere using tesseroids, _Computers & Geosciences_, Volume 96, November 2016, Pages 124-135, ISSN 0098-3004, http://dx.doi.org/10.1016/j.cageo.2016.08.004.
|
||||||
|
|
||||||
|
Article can also be found here http://goo.gl/x9g7gi (researchgate).
|
||||||
|
|
||||||
|
## Usage of _magnetic tesseroids_
|
||||||
|
_Magnetic tesseroids_ are based on the existing program of Leonardo Uieda called tesseroids (Uieda, 2013) of version 1.1 (https://github.com/leouieda/tesseroids/releases/tag/v1.1). It inherits the interface of tesseroids-1.1 but with several changes. Present appendix describes constants and units used be the program, as well as input and output format.
|
||||||
|
### Constants
|
||||||
|
1. Geocentric mean Earth's radius _R_E_ = 6378.137 km.
|
||||||
|
1. Magnetic permeability of a free space _µ_0_ = 4π × 10^-7 H·m^-1
|
||||||
|
|
||||||
|
### List of programs
|
||||||
|
The tessbx, tessby, tessbz are programs that calculate the corresponding components (x - north, y - east, **z - up**) of the magnetic field of the tesseroid model on the computational grid.
|
||||||
|
|
||||||
|
### Input: tesseroid model
|
||||||
|
The input model file should be a text file where each line describe one tesseroid in such space separated format:
|
||||||
|
> `W E S N HEIGHT_OF_TOP HEIGHT_OF_BOTTOM DENSITY SUSCEPTIBILITY BX BY BZ`
|
||||||
|
|
||||||
|
`W`, `E`, `S`, `N` correspond to the western, eastern, southern and northern edges of a tesseroid (_λ_1_, _λ_2_, _ϕ_1_, _ϕ_2_ respectively) expressed in decimal degrees [°].
|
||||||
|
`HEIGHT_OF_TOP` and `HEIGHT_OF_BOTTOM` define the top and the bottom of tesseroid (_r_2_ and _r_1_ respectively). Both are counted from geocentric mean Earth's radius in meters [m]. If a tesseroid should be placed beneath the mean surface, than the values of these parameters should be negative. Note that `HEIGHT_OF_TOP` > `HEIGHT_OF_BOTTOM`.
|
||||||
|
`DENSITY` is the density _ρ_ of tesseroid in kilogram per cubic meter [kg/m^3]
|
||||||
|
`SUSCEPTIBILITY` is the susceptibility _χ_ of tesseroid in SI units.
|
||||||
|
`BX`, `BY` and `BZ` are the components of the magnetizing field in the local North-East-Up Cartesian coordinate system of a tesseroids' geometric center. They can be taken from any core field's model. Values are given in nanotesla [nT].
|
||||||
|
In case of remanent magnetic field modeling, susceptibility must be set 1 SI and `BX`, `BY` and `BZ` values than would define the direction of remanent magnetization vector.
|
||||||
|
This example shows a model made of 3 neighboring tesseroids near the North Pole:
|
||||||
|
> `-74 -73 89 90 -1000.000000 -11650.000000 1.000000 1.000000 334.9504973176 -1969.9308033594 -56572.6324041700`
|
||||||
|
|
||||||
|
> `-73 -72 89 90 -1000.000000 -11650.000000 1.000000 1.000000 370.1879538142 -1968.1093976826 -56571.2826313492`
|
||||||
|
|
||||||
|
> `-72 -71 89 90 -1000.000000 -11650.000000 1.000000 1.000000 405.4388222633 -1965.6409379187 -56569.9502088641`
|
||||||
|
|
||||||
|
### Input: computation grid
|
||||||
|
Computation grid can be regular or irregular and should be also a text file where each line describe the position of one computation point in such space separated format:
|
||||||
|
>`LON LAT ALT`
|
||||||
|
|
||||||
|
`LON` and `LAT` correspond to the longitude and latitude of the point in decimal degrees [°].
|
||||||
|
`ALT` corresponds to the altitude of the point above the mean surface in meters [m].
|
||||||
|
Note that the program tessgrd from original tesseroids-1.1 can be used to create a regular computation grid (see Uieda, 2013).
|
||||||
|
This example shows a grid made of 6 points with the same latitude and the altitude of 400 km:
|
||||||
|
> `-6 51 400000 `
|
||||||
|
|
||||||
|
> `-5.8 51 400000 `
|
||||||
|
|
||||||
|
> `-5.6 51 400000 `
|
||||||
|
|
||||||
|
> `-5.4 51 400000 `
|
||||||
|
|
||||||
|
> `-5.2 51 400000 `
|
||||||
|
|
||||||
|
> `-5 51 400000`
|
||||||
|
|
||||||
|
### Performing calculations
|
||||||
|
Example: to calculate the vertical component of the magnetic field of a model in file modelfile.txt on a grid from file gridpoints.txt one can simply use a console command:
|
||||||
|
```
|
||||||
|
tessbz modelfile.txt < gridpoints.txt > gz_output.txt
|
||||||
|
```
|
||||||
|
|
||||||
|
The result would be written in the file gz_output.txt.
|
||||||
|
### Output format
|
||||||
|
The programs' output is a modified grid file where in the end of each line the calculated value of a corresponding magnetic field component would be written. Values are given in nanotesla [nT] in the local North-East-Up coordinate system of a computational point.
|
||||||
|
### Additional features
|
||||||
|
Magnetic tesseroids support features like piping and integration accuracy adjustment from tesseroids-1.1. Please, check sections in the tesseroids-1.1 manual (Uieda, 2013) relative to the gravity calculation programs to get more information.
|
||||||
|
|
||||||
|
## Utilities
|
||||||
|
### tessutil_magnetize_model
|
||||||
|
This program is made to 'magnetize' any existing tesseroid model by any given main field spherical harmonic model.
|
||||||
|
Usage:
|
||||||
|
```
|
||||||
|
tessutil_magnetize_model [SH coeff file] [input tesseroid model file] [day] [month] [year] [output tesseroid model file]
|
||||||
|
```
|
||||||
|
|
||||||
|
### tessutil_gradient_calculator
|
||||||
|
Gradient calculator (Baykiev et al., in press).
|
||||||
|
Usage:
|
||||||
|
```
|
||||||
|
tessutil_gradient_calculator -bx[Bx grid file] -by[By grid file] -bz[Bx grid file] -o[output component] -c2 >> output_file.dat
|
||||||
|
```
|
||||||
|
|
||||||
|
All grid files should be in tessgrd format. With option `-c1` program reads input grid bz as its direction is upward, with option `-c2` - downward, just as in magnetic tesseroids output. Output of gradient calculator is always in North-East-Down coordinate system.
|
||||||
|
|
||||||
|
Known issue: rounding error when processing grids with spacing equal or less than 0.2 degrees.
|
||||||
|
|
||||||
|
### tessutil_combine_grids
|
||||||
|
Sums calculated grids.
|
||||||
|
Usage:
|
||||||
|
```
|
||||||
|
tessutil_combine_grids [grid file1] [factor1] ... [grid fileN] [factorN] >> output_file.dat
|
||||||
|
```
|
||||||
|
|
||||||
|
Each grid is multiplied by factor (susceptibility) and then the sum of all grids is calculated.
|
||||||
|
|
||||||
|
## Installation (version 1.1)
|
||||||
|
1. Download source code from [GitHub](https://github.com/eldarbaykiev/magnetic-tesseroids):
|
||||||
|
|
||||||
|
```
|
||||||
|
git clone https://github.com/eldarbaykiev/magnetic-tesseroids.git
|
||||||
|
```
|
||||||
|
|
||||||
|
2. On **Linux**, install [OpenBLAS](https://www.openblas.net/) library:
|
||||||
|
|
||||||
|
```
|
||||||
|
sudo apt-get install libopenblas-base libopenblas-dev
|
||||||
|
```
|
||||||
|
|
||||||
|
On **macOS**, make sure that [Xcode](https://developer.apple.com/xcode/) is installed and [Accelerate framework](https://developer.apple.com/documentation/accelerate) is available.
|
||||||
|
|
||||||
|
3. Run **make**
|
||||||
|
|
||||||
|
```
|
||||||
|
make
|
||||||
|
```
|
||||||
|
|
||||||
|
To compile all utilities, run
|
||||||
|
|
||||||
|
```
|
||||||
|
make tools
|
||||||
|
```
|
||||||
74
dep/magnetic_tesseroids/lib/CMakeLists.txt
Normal file
@ -0,0 +1,74 @@
|
|||||||
|
set(CMAKE_C_FLAGS "${CMAKE_C_FLAGS} -O3")
|
||||||
|
set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} -O3")
|
||||||
|
if(${CMAKE_HOST_SYSTEM_NAME} STREQUAL "Windows")
|
||||||
|
# set OpenBLAS directory manually
|
||||||
|
set(OpenBLAS_DIR D:/Library/lib/cmake/openblas)
|
||||||
|
find_library(OpenBLAS REQUIRED)
|
||||||
|
include_directories(${OpenBLAS_INCLUDE_DIRS})
|
||||||
|
elseif(${CMAKE_HOST_SYSTEM_NAME} STREQUAL "Darwin")
|
||||||
|
# set OpenBLAS directory manually
|
||||||
|
set(OpenBLAS_DIR /opt/homebrew/Cellar/openblas/0.3.24/lib/cmake/openblas)
|
||||||
|
find_package(OpenBLAS REQUIRED)
|
||||||
|
include_directories(${OpenBLAS_INCLUDE_DIRS})
|
||||||
|
elseif(${CMAKE_HOST_SYSTEM_NAME} STREQUAL "Linux")
|
||||||
|
set(CMAKE_C_FLAGS "${CMAKE_C_FLAGS} -lm")
|
||||||
|
set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} -lm")
|
||||||
|
endif()
|
||||||
|
|
||||||
|
# 设置库文件的输出地址
|
||||||
|
set(LIBRARY_OUTPUT_PATH ${PROJECT_BINARY_DIR}/lib)
|
||||||
|
|
||||||
|
# 设定库源文件
|
||||||
|
aux_source_directory(. LIBMAGTESS_SRC)
|
||||||
|
|
||||||
|
# 以下部分为库的编译
|
||||||
|
# 注意目标名必须唯一 所以不能直接生成相同名称的动态库与静态库
|
||||||
|
# 注意此处不必为目标名称添加lib前缀和相应后缀,cmake会自行添加
|
||||||
|
add_library(magtess SHARED ${LIBMAGTESS_SRC})
|
||||||
|
# 首先添加静态库的生成命令
|
||||||
|
add_library(magtess_static STATIC ${LIBMAGTESS_SRC})
|
||||||
|
# 设置静态库的输出名称从而获得与动态库名称相同的静态库
|
||||||
|
set_target_properties(magtess_static PROPERTIES OUTPUT_NAME "magtess")
|
||||||
|
# 设置输出目标属性以同时输出动态库与静态库
|
||||||
|
set_target_properties(magtess PROPERTIES CLEAN_DIRECT_OUTPUT 1)
|
||||||
|
set_target_properties(magtess_static PROPERTIES CLEAN_DIRECT_OUTPUT 1)
|
||||||
|
# 设置动态库的版本号
|
||||||
|
set_target_properties(magtess PROPERTIES VERSION ${PROJECT_VERSION} SOVERSION ${PROJECT_VERSION_MAJOR}.${PROJECT_VERSION_MINOR})
|
||||||
|
|
||||||
|
target_link_libraries(magtess PUBLIC ${OpenBLAS_LIBRARIES})
|
||||||
|
target_link_libraries(magtess_static ${OpenBLAS_LIBRARIES})
|
||||||
|
if(${CMAKE_HOST_SYSTEM_NAME} STREQUAL "Linux")
|
||||||
|
target_link_libraries(magtess PUBLIC m)
|
||||||
|
target_link_libraries(magtess PUBLIC openblas)
|
||||||
|
endif()
|
||||||
|
|
||||||
|
set(CONFIG_FILE_PATH lib/cmake/${PROJECT_NAME})
|
||||||
|
|
||||||
|
configure_package_config_file(${PROJECT_SOURCE_DIR}/${PROJECT_NAME}Config.cmake.in
|
||||||
|
${CMAKE_BINARY_DIR}/${PROJECT_NAME}Config.cmake
|
||||||
|
INSTALL_DESTINATION ${CONFIG_FILE_PATH}
|
||||||
|
NO_CHECK_REQUIRED_COMPONENTS_MACRO)
|
||||||
|
|
||||||
|
write_basic_package_version_file(${CMAKE_BINARY_DIR}/${PROJECT_NAME}ConfigVersion.cmake
|
||||||
|
VERSION ${PROJECT_VERSION}
|
||||||
|
COMPATIBILITY SameMajorVersion)
|
||||||
|
|
||||||
|
# 库的安装命令
|
||||||
|
if(WIN32)
|
||||||
|
install(TARGETS magtess DESTINATION lib)
|
||||||
|
install(TARGETS magtess_static DESTINATION lib)
|
||||||
|
else()
|
||||||
|
install(TARGETS magtess magtess_static
|
||||||
|
EXPORT ${PROJECT_NAME}Targets
|
||||||
|
LIBRARY DESTINATION lib
|
||||||
|
ARCHIVE DESTINATION lib)
|
||||||
|
install(EXPORT ${PROJECT_NAME}Targets
|
||||||
|
DESTINATION ${CONFIG_FILE_PATH})
|
||||||
|
install(FILES
|
||||||
|
${CMAKE_BINARY_DIR}/${PROJECT_NAME}Config.cmake
|
||||||
|
${CMAKE_BINARY_DIR}/${PROJECT_NAME}ConfigVersion.cmake
|
||||||
|
DESTINATION ${CONFIG_FILE_PATH})
|
||||||
|
endif()
|
||||||
|
|
||||||
|
file(GLOB LIBMEGTESS_HEAD *.h)
|
||||||
|
install(FILES ${LIBMEGTESS_HEAD} DESTINATION include/magtess)
|
||||||
60
dep/magnetic_tesseroids/lib/constants.h
Executable file
@ -0,0 +1,60 @@
|
|||||||
|
/*
|
||||||
|
Define constants used, like the gravitational constant and unit conversions.
|
||||||
|
|
||||||
|
Values are assigned in file constants.c
|
||||||
|
|
||||||
|
All values are in SI units!
|
||||||
|
*/
|
||||||
|
|
||||||
|
#ifndef _TESSEROIDS_CONSTANTS_H_
|
||||||
|
#define _TESSEROIDS_CONSTANTS_H_
|
||||||
|
|
||||||
|
/* Mean Earth radius [\f$ m \f$] */
|
||||||
|
const double MEAN_EARTH_RADIUS = 6378137.0;
|
||||||
|
const double EARTH_RADIUS_IGRF_KM = 6371.2;
|
||||||
|
|
||||||
|
/* The gravitational constant [\f$ m^3*kg^{-1}*s^{-1} \f$] */
|
||||||
|
const double G = 0.00000000006673;
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
/* Conversion factor from SI units to Eotvos
|
||||||
|
[\f$ \frac{1}{s^2} = 10^9\ Eotvos \f$] */
|
||||||
|
const double SI2EOTVOS = 1000000000.0;
|
||||||
|
const double EOTVOS2SI = 0.000000001;
|
||||||
|
|
||||||
|
/* Conversion factor from SI units to mGal
|
||||||
|
[\f$ 1 \frac{m}{s^2} = 10^5\ mGal \f$] */
|
||||||
|
const double SI2MGAL = 100000.0;
|
||||||
|
|
||||||
|
/* Pi */
|
||||||
|
#ifdef __cplusplus
|
||||||
|
const double PI = 3.1415926535897932384626433832795;
|
||||||
|
#else
|
||||||
|
#define PI 3.1415926535897932384626433832795
|
||||||
|
#endif
|
||||||
|
|
||||||
|
/* minimum distance-to-size ratio for potential computations to be accurate */
|
||||||
|
const double TESSEROID_POT_SIZE_RATIO = 1.5;
|
||||||
|
/* Minimum distance-to-size ratio for gravity computations to be accurate */
|
||||||
|
const double TESSEROID_GX_SIZE_RATIO = 3;
|
||||||
|
const double TESSEROID_GY_SIZE_RATIO = 3;
|
||||||
|
const double TESSEROID_GZ_SIZE_RATIO = 2;
|
||||||
|
/* Minimum distance-to-size ratio for gravity gradient computations to be
|
||||||
|
accurate */
|
||||||
|
const double TESSEROID_GXX_SIZE_RATIO = 3;
|
||||||
|
const double TESSEROID_GXY_SIZE_RATIO = 4.5;
|
||||||
|
const double TESSEROID_GXZ_SIZE_RATIO = 4;
|
||||||
|
const double TESSEROID_GYY_SIZE_RATIO = 3;
|
||||||
|
const double TESSEROID_GYZ_SIZE_RATIO = 4;
|
||||||
|
const double TESSEROID_GZZ_SIZE_RATIO = 3;
|
||||||
|
|
||||||
|
const double M_0 = 4 * (PI) * 0.0000001;
|
||||||
|
|
||||||
|
const double DEG2RAD = (PI)/180.0;
|
||||||
|
|
||||||
|
|
||||||
|
#define FALSE 0
|
||||||
|
#define TRUE 1
|
||||||
|
|
||||||
|
#endif
|
||||||
48
dep/magnetic_tesseroids/lib/geometry.cpp
Executable file
@ -0,0 +1,48 @@
|
|||||||
|
/*
|
||||||
|
Data structures for geometric elements and functions that operate on them.
|
||||||
|
Defines the TESSEROID, SPHERE, and PRISM structures.
|
||||||
|
*/
|
||||||
|
|
||||||
|
|
||||||
|
#include <stdio.h>
|
||||||
|
#include <stdlib.h>
|
||||||
|
#include <math.h>
|
||||||
|
#include <string.h>
|
||||||
|
#include "constants.h"
|
||||||
|
#include "logger.h"
|
||||||
|
#include "geometry.h"
|
||||||
|
|
||||||
|
|
||||||
|
/* Split a tesseroid into 8. */
|
||||||
|
void split_tess(MAG_TESSEROID tess, MAG_TESSEROID *split)
|
||||||
|
{
|
||||||
|
double dlon = 0.5*(tess.e - tess.w),
|
||||||
|
dlat = 0.5*(tess.n - tess.s),
|
||||||
|
dr = 0.5*(tess.r2 - tess.r1),
|
||||||
|
ws[2], ss[2], r1s[2];
|
||||||
|
int i, j, k, t = 0;
|
||||||
|
|
||||||
|
ws[0] = tess.w;
|
||||||
|
ws[1] = tess.w + dlon;
|
||||||
|
ss[0] = tess.s;
|
||||||
|
ss[1] = tess.s + dlat;
|
||||||
|
r1s[0] = tess.r1;
|
||||||
|
r1s[1] = tess.r1 + dr;
|
||||||
|
for(k = 0; k < 2; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < 2; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < 2; i++)
|
||||||
|
{
|
||||||
|
split[t].w = ws[i];
|
||||||
|
split[t].e = ws[i] + dlon;
|
||||||
|
split[t].s = ss[j];
|
||||||
|
split[t].n = ss[j] + dlat;
|
||||||
|
split[t].r1 = r1s[k];
|
||||||
|
split[t].r2 = r1s[k] + dr;
|
||||||
|
split[t].density = tess.density;
|
||||||
|
t++;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
37
dep/magnetic_tesseroids/lib/geometry.h
Executable file
@ -0,0 +1,37 @@
|
|||||||
|
/*
|
||||||
|
Data structures for geometric elements and functions that operate on them.
|
||||||
|
Defines the TESSEROID, SPHERE, and PRISM structures.
|
||||||
|
*/
|
||||||
|
|
||||||
|
#ifndef _MAG_TESSEROIDS_GEOMETRY_H_
|
||||||
|
#define _MAG_TESSEROIDS_GEOMETRY_H_
|
||||||
|
|
||||||
|
|
||||||
|
/* Store information on a tesseroid */
|
||||||
|
typedef struct magtess_struct {
|
||||||
|
/* s, n, w, e in degrees. r1 and r2 are the smaller and larger radius */
|
||||||
|
double density; /* in SI units */
|
||||||
|
double w; /* western longitude border in degrees */
|
||||||
|
double e; /* eastern longitude border in degrees */
|
||||||
|
double s; /* southern latitude border in degrees */
|
||||||
|
double n; /* northern latitude border in degrees */
|
||||||
|
double r1; /* smallest radius border in SI units */
|
||||||
|
double r2; /* largest radius border in SI units */
|
||||||
|
double suscept; /* magnetic susceptibility */
|
||||||
|
double Bx; /* x-component of ambient magnetic field */
|
||||||
|
double By; /* y-component of ambient magnetic field */
|
||||||
|
double Bz; /* z-component of ambient magnetic field */
|
||||||
|
|
||||||
|
double cos_a1;
|
||||||
|
double sin_a1;
|
||||||
|
double cos_b1;
|
||||||
|
double sin_b1;
|
||||||
|
//double Rx;
|
||||||
|
//double Ry;
|
||||||
|
//double Rz;
|
||||||
|
} MAG_TESSEROID;
|
||||||
|
|
||||||
|
|
||||||
|
void split_tess(MAG_TESSEROID tess, MAG_TESSEROID *split);
|
||||||
|
|
||||||
|
#endif
|
||||||
265
dep/magnetic_tesseroids/lib/glq.cpp
Executable file
@ -0,0 +1,265 @@
|
|||||||
|
/*
|
||||||
|
Functions for implementing a Gauss-Legendre Quadrature numerical integration.
|
||||||
|
*/
|
||||||
|
|
||||||
|
|
||||||
|
#include <stdlib.h>
|
||||||
|
#include <math.h>
|
||||||
|
#include "constants.h"
|
||||||
|
#include "logger.h"
|
||||||
|
#include "glq.h"
|
||||||
|
|
||||||
|
|
||||||
|
/* Make a new GLQ structure and set all the parameters needed */
|
||||||
|
GLQ * glq_new(int order, double lower, double upper)
|
||||||
|
{
|
||||||
|
GLQ *glq;
|
||||||
|
int rc;
|
||||||
|
|
||||||
|
glq = (GLQ *)malloc(sizeof(GLQ));
|
||||||
|
if(glq == NULL)
|
||||||
|
{
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
glq->order = order;
|
||||||
|
glq->nodes = (double *)malloc(sizeof(double)*order);
|
||||||
|
if(glq->nodes == NULL)
|
||||||
|
{
|
||||||
|
free(glq);
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
glq->nodes_unscaled = (double *)malloc(sizeof(double)*order);
|
||||||
|
if(glq->nodes_unscaled == NULL)
|
||||||
|
{
|
||||||
|
free(glq);
|
||||||
|
free(glq->nodes);
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
glq->weights = (double *)malloc(sizeof(double)*order);
|
||||||
|
if(glq->weights == NULL)
|
||||||
|
{
|
||||||
|
free(glq);
|
||||||
|
free(glq->nodes);
|
||||||
|
free(glq->nodes_unscaled);
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
rc = glq_nodes(order, glq->nodes_unscaled);
|
||||||
|
if(rc != 0 && rc != 3)
|
||||||
|
{
|
||||||
|
switch(rc)
|
||||||
|
{
|
||||||
|
case 1:
|
||||||
|
log_error("glq_nodes invalid GLQ order %d. Should be >= 2.",
|
||||||
|
order);
|
||||||
|
break;
|
||||||
|
case 2:
|
||||||
|
log_error("glq_nodes NULL pointer for nodes");
|
||||||
|
break;
|
||||||
|
default:
|
||||||
|
log_error("glq_nodes unknown error code %g", rc);
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
glq_free(glq);
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
else if(rc == 3)
|
||||||
|
{
|
||||||
|
log_warning("glq_nodes max iterations reached in root finder");
|
||||||
|
log_warning("nodes might not have desired accuracy %g", GLQ_MAXERROR);
|
||||||
|
}
|
||||||
|
rc = glq_weights(order, glq->nodes_unscaled, glq->weights);
|
||||||
|
if(rc != 0)
|
||||||
|
{
|
||||||
|
switch(rc)
|
||||||
|
{
|
||||||
|
case 1:
|
||||||
|
log_error("glq_weights invalid GLQ order %d. Should be >= 2.",
|
||||||
|
order);
|
||||||
|
break;
|
||||||
|
case 2:
|
||||||
|
log_error("glq_weights NULL pointer for nodes");
|
||||||
|
break;
|
||||||
|
case 3:
|
||||||
|
log_error("glq_weights NULL pointer for weights");
|
||||||
|
break;
|
||||||
|
default:
|
||||||
|
log_error("glq_weights unknown error code %d\n", rc);
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
glq_free(glq);
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
if(glq_set_limits(lower, upper, glq) != 0)
|
||||||
|
{
|
||||||
|
glq_free(glq);
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
return glq;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Free the memory allocated to make a GLQ structure */
|
||||||
|
void glq_free(GLQ *glq)
|
||||||
|
{
|
||||||
|
free(glq->nodes);
|
||||||
|
|
||||||
|
free(glq->nodes_unscaled);
|
||||||
|
free(glq->weights);
|
||||||
|
free(glq);
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Calculates the GLQ nodes using glq_next_root. */
|
||||||
|
int glq_nodes(int order, double *nodes)
|
||||||
|
{
|
||||||
|
register int i;
|
||||||
|
int rc = 0;
|
||||||
|
double initial;
|
||||||
|
|
||||||
|
if(order < 2)
|
||||||
|
{
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
if(nodes == NULL)
|
||||||
|
{
|
||||||
|
return 2;
|
||||||
|
}
|
||||||
|
for(i = 0; i < order; i++)
|
||||||
|
{
|
||||||
|
initial = cos(PI*(order - i - 0.25)/(order + 0.5));
|
||||||
|
if(glq_next_root(initial, i, order, nodes) == 3)
|
||||||
|
{
|
||||||
|
rc = 3;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
return rc;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Put the GLQ nodes to the integration limits IN PLACE. */
|
||||||
|
int glq_set_limits(double lower, double upper, GLQ *glq)
|
||||||
|
{
|
||||||
|
/* Only calculate once to optimize the code */
|
||||||
|
double tmpplus = 0.5*(upper + lower), tmpminus = 0.5*(upper - lower);
|
||||||
|
register int i;
|
||||||
|
|
||||||
|
if(glq->order < 2)
|
||||||
|
{
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
if(glq->nodes == NULL)
|
||||||
|
{
|
||||||
|
return 2;
|
||||||
|
}
|
||||||
|
if(glq->nodes_unscaled == NULL)
|
||||||
|
{
|
||||||
|
return 2;
|
||||||
|
}
|
||||||
|
for(i = 0; i < glq->order; i++)
|
||||||
|
{
|
||||||
|
glq->nodes[i] = tmpminus*glq->nodes_unscaled[i] + tmpplus;
|
||||||
|
|
||||||
|
}
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
/* Calculate the next Legendre polynomial root given the previous root found. */
|
||||||
|
int glq_next_root(double initial, int root_index, int order, double *roots)
|
||||||
|
{
|
||||||
|
double x1, x0, pn, pn_2, pn_1, pn_line, sum;
|
||||||
|
int it = 0;
|
||||||
|
register int n;
|
||||||
|
|
||||||
|
if(order < 2)
|
||||||
|
{
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
if(root_index < 0 || root_index >= order)
|
||||||
|
{
|
||||||
|
return 2;
|
||||||
|
}
|
||||||
|
x1 = initial;
|
||||||
|
do
|
||||||
|
{
|
||||||
|
x0 = x1;
|
||||||
|
|
||||||
|
/* Calculate Pn(x0) */
|
||||||
|
/* Starting from P0(x) and P1(x), */
|
||||||
|
/* find the others using the recursive relation: */
|
||||||
|
/* Pn(x)=(2n-1)xPn_1(x)/n - (n-1)Pn_2(x)/n */
|
||||||
|
pn_1 = 1.; /* This is Po(x) */
|
||||||
|
pn = x0; /* and this P1(x) */
|
||||||
|
for(n = 2; n <= order; n++)
|
||||||
|
{
|
||||||
|
pn_2 = pn_1;
|
||||||
|
pn_1 = pn;
|
||||||
|
pn = ( ((2*n - 1)*x0*pn_1) - ((n - 1)*pn_2) )/n;
|
||||||
|
}
|
||||||
|
/* Now calculate Pn'(x0) using another recursive relation: */
|
||||||
|
/* Pn'(x)=n(xPn(x)-Pn_1(x))/(x*x-1) */
|
||||||
|
pn_line = order*(x0*pn - pn_1)/(x0*x0 - 1);
|
||||||
|
/* Sum the roots found so far */
|
||||||
|
for(n = 0, sum = 0; n < root_index; n++)
|
||||||
|
{
|
||||||
|
sum += 1./(x0 - roots[n]);
|
||||||
|
}
|
||||||
|
/* Update the estimate for the root */
|
||||||
|
x1 = x0 - (double)pn/(pn_line - pn*sum);
|
||||||
|
|
||||||
|
/** Compute the absolute value of x */
|
||||||
|
#define GLQ_ABS(x) ((x) < 0 ? -1*(x) : (x))
|
||||||
|
} while(GLQ_ABS(x1 - x0) > GLQ_MAXERROR && ++it <= GLQ_MAXIT);
|
||||||
|
#undef GLQ_ABS
|
||||||
|
|
||||||
|
roots[root_index] = x1;
|
||||||
|
|
||||||
|
/* Tell the user if stagnation occurred */
|
||||||
|
if(it > GLQ_MAXIT)
|
||||||
|
{
|
||||||
|
return 3;
|
||||||
|
}
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Calculates the weighting coefficients for the GLQ integration. */
|
||||||
|
int glq_weights(int order, double *nodes, double *weights)
|
||||||
|
{
|
||||||
|
register int i, n;
|
||||||
|
double xi, pn, pn_2, pn_1, pn_line;
|
||||||
|
|
||||||
|
if(order < 2)
|
||||||
|
{
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
if(nodes == NULL)
|
||||||
|
{
|
||||||
|
return 2;
|
||||||
|
}
|
||||||
|
if(weights == NULL)
|
||||||
|
{
|
||||||
|
return 3;
|
||||||
|
}
|
||||||
|
for(i = 0; i < order; i++){
|
||||||
|
|
||||||
|
xi = nodes[i];
|
||||||
|
|
||||||
|
/* Find Pn'(xi) with the recursive relation to find Pn and Pn-1: */
|
||||||
|
/* Pn(x)=(2n-1)xPn_1(x)/n - (n-1)Pn_2(x)/n */
|
||||||
|
/* Then use: Pn'(x)=n(xPn(x)-Pn_1(x))/(x*x-1) */
|
||||||
|
|
||||||
|
/* Find Pn and Pn-1 stating from P0 and P1 */
|
||||||
|
pn_1 = 1; /* This is Po(x) */
|
||||||
|
pn = xi; /* and this P1(x) */
|
||||||
|
for(n = 2; n <= order; n++)
|
||||||
|
{
|
||||||
|
pn_2 = pn_1;
|
||||||
|
pn_1 = pn;
|
||||||
|
pn = ((2*n - 1)*xi*pn_1 - (n - 1)*pn_2)/n;
|
||||||
|
}
|
||||||
|
pn_line = order*(xi*pn - pn_1)/(xi*xi - 1.);
|
||||||
|
/* ith weight is: wi = 2/(1 - xi^2)(Pn'(xi)^2) */
|
||||||
|
weights[i] = 2./((1 - xi*xi)*pn_line*pn_line);
|
||||||
|
}
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
182
dep/magnetic_tesseroids/lib/glq.h
Executable file
@ -0,0 +1,182 @@
|
|||||||
|
/*
|
||||||
|
Functions for implementing a Gauss-Legendre Quadrature numerical integration
|
||||||
|
(Hildebrand, 1987).
|
||||||
|
|
||||||
|
Usage example
|
||||||
|
-------------
|
||||||
|
|
||||||
|
To integrate the cossine function from 0 to 90 degrees:
|
||||||
|
|
||||||
|
#include <stdio.h>
|
||||||
|
#include <stdlib.h>
|
||||||
|
#include <math.h>
|
||||||
|
#include "src/c/glq.h"
|
||||||
|
|
||||||
|
int main(){
|
||||||
|
// Create a new glq structure
|
||||||
|
GLQ *glq;
|
||||||
|
double result = 0, a = 0, b = 0.5*3.14;
|
||||||
|
int i;
|
||||||
|
|
||||||
|
glq = glq_new(5, a, b);
|
||||||
|
|
||||||
|
if(glq == NULL){
|
||||||
|
printf("malloc error");
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
|
||||||
|
// Calculate the integral
|
||||||
|
for(i = 0; i < glq->order; i++)
|
||||||
|
result += glq->weights[i]*cos(glq->nodes[i]);
|
||||||
|
|
||||||
|
// Need to multiply by a scale factor of the integration limits
|
||||||
|
result *= 0.5*(b - a);
|
||||||
|
|
||||||
|
printf("Integral of cossine from 0 to 90 degrees = %lf\n", result);
|
||||||
|
|
||||||
|
// Free allocated memory
|
||||||
|
glq_free(glq);
|
||||||
|
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
References
|
||||||
|
----------
|
||||||
|
|
||||||
|
* Hildebrand, F.B (1987): Introduction to numerical analysis.
|
||||||
|
Courier Dover Publications, 2. ed.
|
||||||
|
*/
|
||||||
|
|
||||||
|
#ifndef _TESSEROIDS_GLQ_H_
|
||||||
|
#define _TESSEROIDS_GLQ_H_
|
||||||
|
|
||||||
|
|
||||||
|
/** \var GLQ_MAXIT
|
||||||
|
Max iterations of the root-finder algorithm */
|
||||||
|
const int GLQ_MAXIT = 1000;
|
||||||
|
|
||||||
|
|
||||||
|
/** \var GLQ_MAXERROR
|
||||||
|
Max error allowed for the root-finder algorithm */
|
||||||
|
const double GLQ_MAXERROR = 0.000000000000001;
|
||||||
|
|
||||||
|
|
||||||
|
/** Store the nodes and weights needed for a GLQ integration */
|
||||||
|
typedef struct glq_struct
|
||||||
|
{
|
||||||
|
int order; /**< order of the quadrature, ie number of nodes */
|
||||||
|
double *nodes; /**< abscissas or discretization points of the quadrature */
|
||||||
|
double *weights; /**< weighting coefficients of the quadrature */
|
||||||
|
double *nodes_unscaled; /**< nodes in [-1,1] interval */
|
||||||
|
} GLQ;
|
||||||
|
|
||||||
|
|
||||||
|
/** Make a new GLQ structure and set all the parameters needed
|
||||||
|
|
||||||
|
<b>WARNING</b>: Don't forget to free the memory malloced by this function using
|
||||||
|
glq_free()!
|
||||||
|
|
||||||
|
Prints error and warning messages using the logging.h module.
|
||||||
|
|
||||||
|
@param order order of the quadrature, ie number of nodes
|
||||||
|
@param lower lower integration limit
|
||||||
|
@param upper upper integration limit
|
||||||
|
|
||||||
|
@return GLQ data structure with the nodes and weights calculated. NULL if there
|
||||||
|
was an error with allocation.
|
||||||
|
*/
|
||||||
|
GLQ * glq_new(int order, double lower, double upper);
|
||||||
|
|
||||||
|
|
||||||
|
/** Free the memory allocated to make a GLQ structure
|
||||||
|
|
||||||
|
@param glq pointer to the allocated memory
|
||||||
|
*/
|
||||||
|
void glq_free(GLQ *glq);
|
||||||
|
|
||||||
|
|
||||||
|
/** Put the GLQ nodes to the integration limits <b>IN PLACE</b>.
|
||||||
|
|
||||||
|
Will replace the values of glq.nodes with ones in the specified integration
|
||||||
|
limits.
|
||||||
|
|
||||||
|
In case the GLQ structure was created with glq_new(), the integration limits can
|
||||||
|
be reset using this function.
|
||||||
|
|
||||||
|
@param lower lower integration limit
|
||||||
|
@param upper upper integration limit
|
||||||
|
@param glq pointer to a GLQ structure created with glq_new() and with all
|
||||||
|
necessary memory allocated
|
||||||
|
|
||||||
|
@return Return code:
|
||||||
|
- 0: if everything went OK
|
||||||
|
- 1: if invalid order
|
||||||
|
- 2: if NULL pointer for nodes or nodes_unscaled
|
||||||
|
*/
|
||||||
|
int glq_set_limits(double lower, double upper, GLQ *glq);
|
||||||
|
/** Calculates the GLQ nodes using glq_next_root.
|
||||||
|
|
||||||
|
Nodes will be in the [-1,1] interval. To convert them to the integration limits
|
||||||
|
use glq_scale_nodes
|
||||||
|
|
||||||
|
@param order order of the quadrature, ie how many nodes. Must be >= 2.
|
||||||
|
@param nodes pre-allocated array to return the nodes.
|
||||||
|
|
||||||
|
@return Return code:
|
||||||
|
- 0: if everything went OK
|
||||||
|
- 1: if invalid order
|
||||||
|
- 2: if NULL pointer for nodes
|
||||||
|
- 3: if number of maximum iterations was reached when calculating the root.
|
||||||
|
This usually means that the desired accuracy was not achieved. Default
|
||||||
|
desired accuracy is GLQ_MAXERROR. Default maximum iterations is
|
||||||
|
GLQ_MAXIT.
|
||||||
|
*/
|
||||||
|
int glq_nodes(int order, double *nodes);
|
||||||
|
|
||||||
|
|
||||||
|
/** Calculate the next Legendre polynomial root given the previous root found.
|
||||||
|
|
||||||
|
Uses the root-finder algorithm of:
|
||||||
|
|
||||||
|
Barrera-Figueroa, V., Sosa-Pedroza, J. and López-Bonilla, J., 2006,
|
||||||
|
"Multiple root finder algorithm for Legendre and Chebyshev polynomials via
|
||||||
|
Newton's method", 2006, Annales mathematicae et Informaticae, 33, pp 3-13
|
||||||
|
|
||||||
|
@param initial initial estimate of the next root. I recommend the use of
|
||||||
|
\f$ \cos\left(\pi\frac{(N - i - 0.25)}{N + 0.5}\right) \f$,
|
||||||
|
where \f$ i \f$ is the index of the desired root
|
||||||
|
@param root_index index of the desired root, starting from 0
|
||||||
|
@param order order of the Legendre polynomial, ie number of roots.
|
||||||
|
@param roots array with the roots found so far. Will return the next root in
|
||||||
|
roots[root_index], so make sure to malloc enough space.
|
||||||
|
|
||||||
|
@return Return code:
|
||||||
|
- 0: if everything went OK
|
||||||
|
- 1: if order is not valid
|
||||||
|
- 2: if root_index is not valid (negative)
|
||||||
|
- 3: if number of maximum iterations was reached when calculating the root.
|
||||||
|
This usually means that the desired accuracy was not achieved. Default
|
||||||
|
desired accuracy is GLQ_MAXERROR. Default maximum iterations is
|
||||||
|
GLQ_MAXIT.
|
||||||
|
*/
|
||||||
|
int glq_next_root(double initial, int root_index, int order,
|
||||||
|
double *roots);
|
||||||
|
|
||||||
|
|
||||||
|
/** Calculates the weighting coefficients for the GLQ integration.
|
||||||
|
|
||||||
|
@param order order of the quadrature, ie number of nodes and weights.
|
||||||
|
@param nodes array containing the GLQ nodes calculated by glq_nodes.
|
||||||
|
<b>IMPORTANT</b>: needs the nodes in [-1,1] interval! Scaled nodes
|
||||||
|
will result in wrong weights!
|
||||||
|
@param weights pre-allocated array to return the weights
|
||||||
|
|
||||||
|
@return Return code:
|
||||||
|
- 0: if everything went OK
|
||||||
|
- 1: if order is not valid
|
||||||
|
- 2: if nodes is a NULL pointer
|
||||||
|
- 3: if weights is a NULL pointer
|
||||||
|
*/
|
||||||
|
int glq_weights(int order, double *nodes, double *weights);
|
||||||
|
|
||||||
|
#endif
|
||||||
628
dep/magnetic_tesseroids/lib/grav_tess.cpp
Executable file
@ -0,0 +1,628 @@
|
|||||||
|
/*
|
||||||
|
Functions that calculate the gravitational potential and its first and second
|
||||||
|
derivatives for the tesseroid.
|
||||||
|
|
||||||
|
References
|
||||||
|
----------
|
||||||
|
|
||||||
|
* Grombein, T.; Seitz, K.; Heck, B. (2010): Untersuchungen zur effizienten
|
||||||
|
Berechnung topographischer Effekte auf den Gradiententensor am Fallbeispiel der
|
||||||
|
Satellitengradiometriemission GOCE.
|
||||||
|
KIT Scientific Reports 7547, ISBN 978-3-86644-510-9, KIT Scientific Publishing,
|
||||||
|
Karlsruhe, Germany.
|
||||||
|
*/
|
||||||
|
|
||||||
|
|
||||||
|
#include <math.h>
|
||||||
|
#include "logger.h"
|
||||||
|
#include "geometry.h"
|
||||||
|
#include "glq.h"
|
||||||
|
#include "constants.h"
|
||||||
|
#include "grav_tess.h"
|
||||||
|
|
||||||
|
|
||||||
|
/* Calculates the field of a tesseroid model at a given point. */
|
||||||
|
double calc_tess_model(MAG_TESSEROID *model, int size, double lonp, double latp, double rp, GLQ *glq_lon, GLQ *glq_lat, GLQ *glq_r, double (*field)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ))
|
||||||
|
{
|
||||||
|
double res;
|
||||||
|
int tess;
|
||||||
|
|
||||||
|
res = 0;
|
||||||
|
for(tess = 0; tess < size; tess++)
|
||||||
|
{
|
||||||
|
if(lonp >= model[tess].w && lonp <= model[tess].e &&
|
||||||
|
latp >= model[tess].s && latp <= model[tess].n &&
|
||||||
|
rp >= model[tess].r1 && rp <= model[tess].r2)
|
||||||
|
{
|
||||||
|
log_warning("Point (%g %g %g) is on tesseroid %d: %g %g %g %g %g %g %g. Can't guarantee accuracy.",
|
||||||
|
lonp, latp, rp - MEAN_EARTH_RADIUS, tess,
|
||||||
|
model[tess].w, model[tess].e, model[tess].s,
|
||||||
|
model[tess].n, model[tess].r2 - MEAN_EARTH_RADIUS,
|
||||||
|
model[tess].r1 - MEAN_EARTH_RADIUS,
|
||||||
|
model[tess].density);
|
||||||
|
}
|
||||||
|
glq_set_limits(model[tess].w, model[tess].e, glq_lon);
|
||||||
|
glq_set_limits(model[tess].s, model[tess].n, glq_lat);
|
||||||
|
glq_set_limits(model[tess].r1, model[tess].r2, glq_r);
|
||||||
|
res += field(model[tess], lonp, latp, rp, *glq_lon, *glq_lat, *glq_r);
|
||||||
|
}
|
||||||
|
return res;
|
||||||
|
}
|
||||||
|
|
||||||
|
void calc_tess_model_triple(MAG_TESSEROID *model, int size, double lonp, double latp, double rp, GLQ *glq_lon, GLQ *glq_lat, GLQ *glq_r, void (*field_triple)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ, double*), double *res)
|
||||||
|
{
|
||||||
|
double r1, r2, r3, ri[3];
|
||||||
|
int tess;
|
||||||
|
|
||||||
|
res[0] = 0;
|
||||||
|
res[1] = 0;
|
||||||
|
res[2] = 0;
|
||||||
|
for(tess = 0; tess < size; tess++)
|
||||||
|
{
|
||||||
|
ri[0] = 0;
|
||||||
|
ri[1] = 0;
|
||||||
|
ri[2] = 0;
|
||||||
|
|
||||||
|
if(lonp >= model[tess].w && lonp <= model[tess].e &&
|
||||||
|
latp >= model[tess].s && latp <= model[tess].n &&
|
||||||
|
rp >= model[tess].r1 && rp <= model[tess].r2)
|
||||||
|
{
|
||||||
|
log_warning("Point (%g %g %g) is on tesseroid %d: %g %g %g %g %g %g %g. Can't guarantee accuracy.",
|
||||||
|
lonp, latp, rp - MEAN_EARTH_RADIUS, tess,
|
||||||
|
model[tess].w, model[tess].e, model[tess].s,
|
||||||
|
model[tess].n, model[tess].r2 - MEAN_EARTH_RADIUS,
|
||||||
|
model[tess].r1 - MEAN_EARTH_RADIUS,
|
||||||
|
model[tess].density);
|
||||||
|
}
|
||||||
|
glq_set_limits(model[tess].w, model[tess].e, glq_lon);
|
||||||
|
glq_set_limits(model[tess].s, model[tess].n, glq_lat);
|
||||||
|
glq_set_limits(model[tess].r1, model[tess].r2, glq_r);
|
||||||
|
field_triple(model[tess], lonp, latp, rp, *glq_lon, *glq_lat, *glq_r, ri);
|
||||||
|
|
||||||
|
res[0] += ri[0];
|
||||||
|
res[1] += ri[1];
|
||||||
|
res[2] += ri[2];
|
||||||
|
}
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
/* Adaptatively calculate the field of a tesseroid model at a given point */
|
||||||
|
double calc_tess_model_adapt(MAG_TESSEROID *model, int size, double lonp, double latp, double rp, GLQ *glq_lon, GLQ *glq_lat, GLQ *glq_r, double (*field)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ), double ratio)
|
||||||
|
{
|
||||||
|
double res, dist, lont, latt, rt, d2r = PI/180.;
|
||||||
|
int tess;
|
||||||
|
MAG_TESSEROID split[8];
|
||||||
|
|
||||||
|
res = 0;
|
||||||
|
for(tess = 0; tess < size; tess++)
|
||||||
|
{
|
||||||
|
rt = model[tess].r2;
|
||||||
|
lont = 0.5*(model[tess].w + model[tess].e);
|
||||||
|
latt = 0.5*(model[tess].s + model[tess].n);
|
||||||
|
dist = sqrt(rp*rp + rt*rt - 2*rp*rt*(sin(d2r*latp)*sin(d2r*latt) +
|
||||||
|
cos(d2r*latp)*cos(d2r*latt)*cos(d2r*(lonp - lont))));
|
||||||
|
|
||||||
|
/* Would get stuck in infinite loop if dist = 0 and get wrong results if
|
||||||
|
inside de tesseroid. Still do the calculation but warn user that it's
|
||||||
|
probably wrong. */
|
||||||
|
if(lonp >= model[tess].w && lonp <= model[tess].e &&
|
||||||
|
latp >= model[tess].s && latp <= model[tess].n &&
|
||||||
|
rp >= model[tess].r1 && rp <= model[tess].r2)
|
||||||
|
{
|
||||||
|
log_warning("Point (%g %g %g) is on top of tesseroid %d: %g %g %g %g %g %g %g. Can't guarantee accuracy.",
|
||||||
|
lonp, latp, rp - MEAN_EARTH_RADIUS, tess,
|
||||||
|
model[tess].w, model[tess].e, model[tess].s,
|
||||||
|
model[tess].n, model[tess].r2 - MEAN_EARTH_RADIUS,
|
||||||
|
model[tess].r1 - MEAN_EARTH_RADIUS,
|
||||||
|
model[tess].density);
|
||||||
|
glq_set_limits(model[tess].w, model[tess].e, glq_lon);
|
||||||
|
glq_set_limits(model[tess].s, model[tess].n, glq_lat);
|
||||||
|
glq_set_limits(model[tess].r1, model[tess].r2, glq_r);
|
||||||
|
res += field(model[tess], lonp, latp, rp, *glq_lon, *glq_lat,
|
||||||
|
*glq_r);
|
||||||
|
}
|
||||||
|
/* Check if the computation point is at an acceptable distance. If not
|
||||||
|
split the tesseroid using the given ratio */
|
||||||
|
else if(
|
||||||
|
dist < ratio*MEAN_EARTH_RADIUS*d2r*(model[tess].e - model[tess].w) ||
|
||||||
|
dist < ratio*MEAN_EARTH_RADIUS*d2r*(model[tess].n - model[tess].s) ||
|
||||||
|
dist < ratio*(model[tess].r2 - model[tess].r1))
|
||||||
|
{
|
||||||
|
log_debug("Splitting tesseroid %d (%g %g %g %g %g %g %g) at point (%g %g %g) using ratio %g",
|
||||||
|
tess, model[tess].w, model[tess].e, model[tess].s,
|
||||||
|
model[tess].n, model[tess].r2 - MEAN_EARTH_RADIUS,
|
||||||
|
model[tess].r1 - MEAN_EARTH_RADIUS, model[tess].density,
|
||||||
|
lonp, latp, rp - MEAN_EARTH_RADIUS, ratio);
|
||||||
|
/* Do it recursively until ratio*size is smaller than distance */
|
||||||
|
split_tess(model[tess], split);
|
||||||
|
res += calc_tess_model_adapt(split, 8, lonp, latp, rp, glq_lon,
|
||||||
|
glq_lat, glq_r, field, ratio);
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
glq_set_limits(model[tess].w, model[tess].e, glq_lon);
|
||||||
|
glq_set_limits(model[tess].s, model[tess].n, glq_lat);
|
||||||
|
glq_set_limits(model[tess].r1, model[tess].r2, glq_r);
|
||||||
|
res += field(model[tess], lonp, latp, rp, *glq_lon, *glq_lat,
|
||||||
|
*glq_r);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
return res;
|
||||||
|
}
|
||||||
|
|
||||||
|
/* Calculates gxx caused by a tesseroid. */
|
||||||
|
double tess_gxx(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, kphi, coslatp, coslatc, sinlatp, sinlatc,
|
||||||
|
coslon, rc, kappa, res;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*(sinlatp*sinlatc +
|
||||||
|
coslatp*coslatc*coslon);
|
||||||
|
|
||||||
|
kphi = coslatp*sinlatc - sinlatp*coslatc*coslon;
|
||||||
|
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
res += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*rc*kphi*rc*kphi - l_sqr)/pow(l_sqr, 2.5);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
return res;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Calculates gxy caused by a tesseroid. */
|
||||||
|
double tess_gxy(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, kphi, coslatp, coslatc, sinlatp, sinlatc,
|
||||||
|
coslon, sinlon, rc, kappa, deltax, deltay, res;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
sinlon = sin(d2r*(glq_lon.nodes[k] - lonp));
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*(sinlatp*sinlatc +
|
||||||
|
coslatp*coslatc*coslon);
|
||||||
|
|
||||||
|
kphi = coslatp*sinlatc - sinlatp*coslatc*coslon;
|
||||||
|
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
deltax = rc*kphi;
|
||||||
|
|
||||||
|
deltay = rc*coslatc*sinlon;
|
||||||
|
|
||||||
|
res += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltax*deltay)/pow(l_sqr, 2.5);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
return res;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Calculates gxz caused by a tesseroid. */
|
||||||
|
double tess_gxz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, kphi, coslatp, coslatc, sinlatp, sinlatc,
|
||||||
|
coslon, cospsi, rc, kappa, deltax, deltaz, res;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
|
||||||
|
cospsi = sinlatp*sinlatc + coslatp*coslatc*coslon;
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*cospsi;
|
||||||
|
|
||||||
|
kphi = coslatp*sinlatc - sinlatp*coslatc*coslon;
|
||||||
|
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
deltax = rc*kphi;
|
||||||
|
|
||||||
|
deltaz = rc*cospsi - rp;
|
||||||
|
|
||||||
|
res += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltax*deltaz)/pow(l_sqr, 2.5);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
return res;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Calculates gyy caused by a tesseroid. */
|
||||||
|
double tess_gyy(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, coslatp, coslatc, sinlatp, sinlatc,
|
||||||
|
coslon, sinlon, rc, kappa, deltay, res;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
sinlon = sin(d2r*(glq_lon.nodes[k] - lonp));
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*(sinlatp*sinlatc +
|
||||||
|
coslatp*coslatc*coslon);
|
||||||
|
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
deltay = rc*coslatc*sinlon;
|
||||||
|
|
||||||
|
res += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltay*deltay - l_sqr)/pow(l_sqr, 2.5);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
return res;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Calculates gyz caused by a tesseroid. */
|
||||||
|
double tess_gyz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, coslatp, coslatc, sinlatp, sinlatc,
|
||||||
|
coslon, sinlon, cospsi, rc, kappa, deltay, deltaz, res;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
sinlon = sin(d2r*(glq_lon.nodes[k] - lonp));
|
||||||
|
|
||||||
|
cospsi = sinlatp*sinlatc + coslatp*coslatc*coslon;
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*cospsi;
|
||||||
|
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
deltay = rc*coslatc*sinlon;
|
||||||
|
|
||||||
|
deltaz = rc*cospsi - rp;
|
||||||
|
|
||||||
|
res += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltay*deltaz)/pow(l_sqr, 2.5);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
return res;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Calculates gzz caused by a tesseroid. */
|
||||||
|
double tess_gzz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, coslatp, coslatc, sinlatp, sinlatc,
|
||||||
|
coslon, cospsi, rc, kappa, deltaz, res;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
|
||||||
|
cospsi = sinlatp*sinlatc + coslatp*coslatc*coslon;
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*cospsi;
|
||||||
|
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
deltaz = rc*cospsi - rp;
|
||||||
|
|
||||||
|
res += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltaz*deltaz - l_sqr)/pow(l_sqr, 2.5);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
return res;
|
||||||
|
}
|
||||||
|
|
||||||
|
/*Calculate three gravity gradient components simultaneously*/
|
||||||
|
void tess_gxz_gyz_gzz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r, double *res)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, coslatp, coslatc, sinlatp, sinlatc, sinlon,
|
||||||
|
coslon, cospsi, rc, kappa, deltaz, deltax, deltay, kphi,
|
||||||
|
res_gxz, res_gyz, res_gzz;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res_gxz = 0;
|
||||||
|
res_gyz = 0;
|
||||||
|
res_gzz = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
sinlon = sin(d2r*(glq_lon.nodes[k] - lonp));
|
||||||
|
cospsi = sinlatp*sinlatc + coslatp*coslatc*coslon;
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*cospsi;
|
||||||
|
kphi = coslatp*sinlatc - sinlatp*coslatc*coslon;
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
deltax = rc*kphi;
|
||||||
|
deltay = rc*coslatc*sinlon;
|
||||||
|
deltaz = rc*cospsi - rp;
|
||||||
|
|
||||||
|
res_gxz += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltax*deltaz)/pow(l_sqr, 2.5);
|
||||||
|
|
||||||
|
res_gyz += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltay*deltaz)/pow(l_sqr, 2.5);
|
||||||
|
|
||||||
|
res_gzz += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltaz*deltaz - l_sqr)/pow(l_sqr, 2.5);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res_gxz *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res_gyz *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res_gzz *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res[0] = res_gxz;
|
||||||
|
res[1] = res_gyz;
|
||||||
|
res[2] = res_gzz;
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
void tess_gxx_gxy_gxz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r, double *res)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, coslatp, coslatc, sinlatp, sinlatc, sinlon,
|
||||||
|
coslon, cospsi, rc, kappa, deltaz, deltax, deltay, kphi,
|
||||||
|
res_gxx, res_gxy, res_gxz;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res_gxx = 0;
|
||||||
|
res_gxy = 0;
|
||||||
|
res_gxz = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
sinlon = sin(d2r*(glq_lon.nodes[k] - lonp));
|
||||||
|
cospsi = sinlatp*sinlatc + coslatp*coslatc*coslon;
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*cospsi;
|
||||||
|
kphi = coslatp*sinlatc - sinlatp*coslatc*coslon;
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
deltax = rc*kphi;
|
||||||
|
deltay = rc*coslatc*sinlon;
|
||||||
|
deltaz = rc*cospsi - rp;
|
||||||
|
|
||||||
|
res_gxx += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*rc*kphi*rc*kphi - l_sqr)/pow(l_sqr, 2.5);
|
||||||
|
|
||||||
|
res_gxy += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltax*deltay)/pow(l_sqr, 2.5);
|
||||||
|
|
||||||
|
res_gxz += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltax*deltaz)/pow(l_sqr, 2.5);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res_gxx *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res_gxy *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res_gxz *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res[0] = res_gxx;
|
||||||
|
res[1] = res_gxy;
|
||||||
|
res[2] = res_gxz;
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
void tess_gxy_gyy_gyz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon,
|
||||||
|
GLQ glq_lat, GLQ glq_r, double *res)
|
||||||
|
{
|
||||||
|
double d2r = PI/180., l_sqr, coslatp, coslatc, sinlatp, sinlatc, sinlon,
|
||||||
|
coslon, cospsi, rc, kappa, deltaz, deltax, deltay, kphi,
|
||||||
|
res_gxy, res_gyy, res_gyz;
|
||||||
|
register int i, j, k;
|
||||||
|
|
||||||
|
coslatp = cos(d2r*latp);
|
||||||
|
sinlatp = sin(d2r*latp);
|
||||||
|
|
||||||
|
res_gxy = 0;
|
||||||
|
res_gyy = 0;
|
||||||
|
res_gyz = 0;
|
||||||
|
|
||||||
|
for(k = 0; k < glq_lon.order; k++)
|
||||||
|
{
|
||||||
|
for(j = 0; j < glq_lat.order; j++)
|
||||||
|
{
|
||||||
|
for(i = 0; i < glq_r.order; i++)
|
||||||
|
{
|
||||||
|
rc = glq_r.nodes[i];
|
||||||
|
sinlatc = sin(d2r*glq_lat.nodes[j]);
|
||||||
|
coslatc = cos(d2r*glq_lat.nodes[j]);
|
||||||
|
coslon = cos(d2r*(lonp - glq_lon.nodes[k]));
|
||||||
|
sinlon = sin(d2r*(glq_lon.nodes[k] - lonp));
|
||||||
|
cospsi = sinlatp*sinlatc + coslatp*coslatc*coslon;
|
||||||
|
|
||||||
|
l_sqr = rp*rp + rc*rc - 2*rp*rc*cospsi;
|
||||||
|
kphi = coslatp*sinlatc - sinlatp*coslatc*coslon;
|
||||||
|
kappa = rc*rc*coslatc;
|
||||||
|
|
||||||
|
deltax = rc*kphi;
|
||||||
|
deltay = rc*coslatc*sinlon;
|
||||||
|
deltaz = rc*cospsi - rp;
|
||||||
|
|
||||||
|
res_gxy += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltax*deltay)/pow(l_sqr, 2.5);
|
||||||
|
|
||||||
|
res_gyy += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltay*deltay - l_sqr)/pow(l_sqr, 2.5);
|
||||||
|
|
||||||
|
res_gyz += glq_lon.weights[k]*glq_lat.weights[j]*glq_r.weights[i]*
|
||||||
|
kappa*(3*deltay*deltaz)/pow(l_sqr, 2.5);
|
||||||
|
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
res_gxy *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res_gyy *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res_gyz *= SI2EOTVOS*G*tess.density*d2r*(tess.e - tess.w)*d2r*(tess.n - tess.s)*
|
||||||
|
(tess.r2 - tess.r1)*0.125;
|
||||||
|
|
||||||
|
res[0] = res_gxy;
|
||||||
|
res[1] = res_gyy;
|
||||||
|
res[2] = res_gyz;
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
93
dep/magnetic_tesseroids/lib/grav_tess.h
Executable file
@ -0,0 +1,93 @@
|
|||||||
|
/*
|
||||||
|
Functions that calculate the gravitational potential and its first and second
|
||||||
|
derivatives for the MAG_tesseroid.
|
||||||
|
|
||||||
|
The gravity gradients can be calculated using the general formula of
|
||||||
|
Grombein et al. (2010).
|
||||||
|
The integrals are solved using the Gauss-Legendre Quadrature rule
|
||||||
|
(Asgharzadeh et al., 2007).
|
||||||
|
|
||||||
|
The derivatives of the potential are made with respect to the local coordinate
|
||||||
|
system x->North, y->East, z->Up (away from center of the Earth).
|
||||||
|
|
||||||
|
To maintain the standard convention, only for component gz the z axis is
|
||||||
|
inverted, so a positive density results in positive gz.
|
||||||
|
|
||||||
|
Example
|
||||||
|
-------
|
||||||
|
|
||||||
|
To calculate the gzz component due to a MAG_tesseroid on a regular grid:
|
||||||
|
|
||||||
|
#include <stdio.h>
|
||||||
|
#include "glq.h"r
|
||||||
|
#include "constants.h"
|
||||||
|
#include "grav_tess.h"
|
||||||
|
|
||||||
|
int main()
|
||||||
|
{
|
||||||
|
MAG_TESSEROID tess = {1000, 44, 46, -1, 1, MEAN_EARTH_RADIUS - 100000,
|
||||||
|
MEAN_EARTH_RADIUS};
|
||||||
|
GLQ *glqlon, *glqlat, *glqr;
|
||||||
|
double lon, lat, r = MEAN_EARTH_RADIUS + 1500000, res;
|
||||||
|
int order = 8;
|
||||||
|
|
||||||
|
glqlon = glq_new(order, tess.w, tess.e);
|
||||||
|
glqlat = glq_new(order, tess.s, tess.n);
|
||||||
|
glqr = glq_new(order, tess.r1, tess.r2);
|
||||||
|
|
||||||
|
for(lat = 20; lat <= 70; lat += 0.5)
|
||||||
|
{
|
||||||
|
for(lon = -25; lon <= 25; lon += 0.5)
|
||||||
|
{
|
||||||
|
res = tess_gzz(tess, lon, lat, r, *glqlon, *glqlat, *glqr);
|
||||||
|
printf("%g %g %g\n", lon, lat, res);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
glq_free(glqlon);
|
||||||
|
glq_free(glqlat);
|
||||||
|
glq_free(glqr);
|
||||||
|
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
References
|
||||||
|
----------
|
||||||
|
|
||||||
|
Asgharzadeh, M.F., von Frese, R.R.B., Kim, H.R., Leftwich, T.E. & Kim, J.W.
|
||||||
|
(2007): Spherical prism gravity effects by Gauss-Legendre quadrature integration.
|
||||||
|
Geophysical Journal International, 169, 1-11.
|
||||||
|
|
||||||
|
Grombein, T.; Seitz, K.; Heck, B. (2010): Untersuchungen zur effizienten
|
||||||
|
Berechnung topographischer Effekte auf den Gradiententensor am Fallbeispiel der
|
||||||
|
Satellitengradiometriemission GOCE.
|
||||||
|
KIT Scientific Reports 7547, ISBN 978-3-86644-510-9, KIT Scientific Publishing,
|
||||||
|
Karlsruhe, Germany.
|
||||||
|
*/
|
||||||
|
|
||||||
|
#ifndef _MAG_TESSEROIDS_GRAV_TESS_H_
|
||||||
|
#define _MAG_TESSEROIDS_GRAV_TESS_H_
|
||||||
|
|
||||||
|
|
||||||
|
/* Needed for definition of MAG_TESSEROID */
|
||||||
|
#include "geometry.h"
|
||||||
|
/* Needed for definition of GLQ */
|
||||||
|
#include "glq.h"
|
||||||
|
|
||||||
|
double calc_tess_model(MAG_TESSEROID *model, int size, double lonp, double latp, double rp, GLQ *glq_lon, GLQ *glq_lat, GLQ *glq_r, double (*field)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ));
|
||||||
|
void calc_tess_model_triple(MAG_TESSEROID *model, int size, double lonp, double latp, double rp, GLQ *glq_lon, GLQ *glq_lat, GLQ *glq_r,
|
||||||
|
void (*field_triple)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ, double*), double *res);
|
||||||
|
double calc_tess_model_adapt(MAG_TESSEROID *model, int size, double lonp, double latp, double rp, GLQ *glq_lon, GLQ *glq_lat, GLQ *glq_r, double (*field)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ), double ratio);
|
||||||
|
|
||||||
|
double tess_gxx(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r);
|
||||||
|
double tess_gxy(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r);
|
||||||
|
double tess_gxz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r);
|
||||||
|
double tess_gyy(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r);
|
||||||
|
double tess_gyz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r);
|
||||||
|
double tess_gzz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r);
|
||||||
|
|
||||||
|
void tess_gxz_gyz_gzz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r, double *res);
|
||||||
|
void tess_gxx_gxy_gxz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r, double *res);
|
||||||
|
void tess_gxy_gyy_gyz(MAG_TESSEROID tess, double lonp, double latp, double rp, GLQ glq_lon, GLQ glq_lat, GLQ glq_r, double *res);
|
||||||
|
|
||||||
|
#endif
|
||||||
144
dep/magnetic_tesseroids/lib/linalg.cpp
Executable file
@ -0,0 +1,144 @@
|
|||||||
|
/*
|
||||||
|
Functions matrix and vector multiplications.
|
||||||
|
|
||||||
|
*/
|
||||||
|
|
||||||
|
#include "linalg.h"
|
||||||
|
#include "constants.h"
|
||||||
|
#include <math.h>
|
||||||
|
|
||||||
|
//macOS only!
|
||||||
|
//#include <Accelerate/Accelerate.h>
|
||||||
|
|
||||||
|
#ifdef __linux__ // Debian, Ubuntu, Gentoo, Fedora, openSUSE, RedHat, Centos and other
|
||||||
|
#include <cblas.h>
|
||||||
|
#elif _WINDOWS || __WIN32__ // Added for windows by Yi Zhang on 2021-08-26
|
||||||
|
#include <cblas.h>
|
||||||
|
#elif defined(__APPLE__) && defined(__MACH__)
|
||||||
|
#include <Accelerate/Accelerate.h>
|
||||||
|
#else
|
||||||
|
|
||||||
|
#endif
|
||||||
|
|
||||||
|
/* Calculate magnetization vector in a coordinate system of a given point */
|
||||||
|
void conv_vect_cblas(double *vect, double lon1, double lat1, double lon2, double lat2, double *res)
|
||||||
|
{
|
||||||
|
double a1 = DEG2RAD*lat1;
|
||||||
|
double b1 = DEG2RAD*lon1;
|
||||||
|
double a2 = DEG2RAD*lat2;
|
||||||
|
double b2 = DEG2RAD*lon2;
|
||||||
|
|
||||||
|
double cos_a1 = cos(PI/2.0-a1);
|
||||||
|
double sin_a1 = sin(PI/2.0-a1);
|
||||||
|
double cos_a2 = cos(PI/2.0-a2);
|
||||||
|
double sin_a2 = sin(PI/2.0-a2);
|
||||||
|
|
||||||
|
double cos_b1 = cos(b1);
|
||||||
|
double sin_b1 = sin(b1);
|
||||||
|
double cos_b2 = cos(b2);
|
||||||
|
double sin_b2 = sin(b2);
|
||||||
|
|
||||||
|
double Z1Y1[9] = {cos_a1*cos_b1, -sin_b1, cos_b1*sin_a1, cos_a1*sin_b1, cos_b1, sin_a1*sin_b1, -sin_a1, 0, cos_a1};
|
||||||
|
double Z2Y2t[9] = {-cos_a2*cos_b2, -cos_a2*sin_b2, sin_a2, -sin_b2, cos_b2, 0, cos_b2*sin_a2, sin_a2*sin_b2, cos_a2};
|
||||||
|
double R[9];
|
||||||
|
|
||||||
|
cblas_dgemm(CblasColMajor, CblasNoTrans, CblasNoTrans, 3, 3, 3, 1.0, Z1Y1, 3, Z2Y2t, 3, 0.0, R, 3);
|
||||||
|
|
||||||
|
R[0] = -R[0];
|
||||||
|
R[3] = -R[3];
|
||||||
|
R[6] = -R[6];
|
||||||
|
|
||||||
|
cblas_dgemv(CblasRowMajor, CblasNoTrans, 3, 3, 1.0, R, 3, vect, 1, 0.0, res, 1);
|
||||||
|
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
void conv_vect_cblas_precalc(double *vect, double cos_a1, double sin_a1, double cos_b1, double sin_b1, double cos_a2, double sin_a2, double cos_b2, double sin_b2, double *res)
|
||||||
|
{
|
||||||
|
// double a1 = DEG2RAD*lat1;
|
||||||
|
// double b1 = DEG2RAD*lon1;
|
||||||
|
// double a2 = DEG2RAD*lat2;
|
||||||
|
// double b2 = DEG2RAD*lon2;
|
||||||
|
|
||||||
|
// double cos_a1 = cos(PI/2.0-a1);
|
||||||
|
// double sin_a1 = sin(PI/2.0-a1);
|
||||||
|
// double cos_a2 = cos(PI/2.0-a2);
|
||||||
|
// double sin_a2 = sin(PI/2.0-a2);
|
||||||
|
|
||||||
|
// double cos_b1 = cos(b1);
|
||||||
|
// double sin_b1 = sin(b1);
|
||||||
|
// double cos_b2 = cos(b2);
|
||||||
|
// double sin_b2 = sin(b2);
|
||||||
|
|
||||||
|
double Z1Y1[9] = {cos_a1*cos_b1, -sin_b1, cos_b1*sin_a1, cos_a1*sin_b1, cos_b1, sin_a1*sin_b1, -sin_a1, 0, cos_a1};
|
||||||
|
double Z2Y2t[9] = {-cos_a2*cos_b2, -cos_a2*sin_b2, sin_a2, -sin_b2, cos_b2, 0, cos_b2*sin_a2, sin_a2*sin_b2, cos_a2};
|
||||||
|
double R[9];
|
||||||
|
|
||||||
|
cblas_dgemm(CblasColMajor, CblasNoTrans, CblasNoTrans, 3, 3, 3, 1.0, Z1Y1, 3, Z2Y2t, 3, 0.0, R, 3);
|
||||||
|
|
||||||
|
R[0] = -R[0];
|
||||||
|
R[3] = -R[3];
|
||||||
|
R[6] = -R[6];
|
||||||
|
|
||||||
|
cblas_dgemv(CblasRowMajor, CblasNoTrans, 3, 3, 1.0, R, 3, vect, 1, 0.0, res, 1);
|
||||||
|
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
void from_loc_sphr_to_cart(double* columnvect_xyzloc, double colatitude, double longitude, double* columnvect_res)
|
||||||
|
{
|
||||||
|
/*IMPORTANT: this subroutine is in the coordinate system NED*/
|
||||||
|
|
||||||
|
double phi = colatitude*DEG2RAD;
|
||||||
|
double lambda = longitude*DEG2RAD;
|
||||||
|
|
||||||
|
double cos_phi = cos(phi);
|
||||||
|
double sin_phi = sin(phi);
|
||||||
|
double cos_lambda = cos(lambda);
|
||||||
|
double sin_lambda = sin(lambda);
|
||||||
|
|
||||||
|
double columnvect_phi_unit[3] = {-sin_phi*cos_lambda, -sin_phi*sin_lambda, cos_phi};
|
||||||
|
double columnvect_lambda_unit[3] = {-sin_lambda, cos_lambda, 0};
|
||||||
|
double columnvect_r_unit[3] = {cos_phi*cos_lambda, cos_phi*sin_lambda, sin_phi};
|
||||||
|
|
||||||
|
columnvect_res[0] = columnvect_phi_unit[0]*columnvect_xyzloc[0]+columnvect_lambda_unit[0]*columnvect_xyzloc[1]+columnvect_r_unit[0]*columnvect_xyzloc[2];
|
||||||
|
columnvect_res[1] = columnvect_phi_unit[1]*columnvect_xyzloc[0]+columnvect_lambda_unit[1]*columnvect_xyzloc[1]+columnvect_r_unit[1]*columnvect_xyzloc[2];
|
||||||
|
columnvect_res[2] = columnvect_phi_unit[2]*columnvect_xyzloc[0]+columnvect_lambda_unit[2]*columnvect_xyzloc[1]+columnvect_r_unit[2]*columnvect_xyzloc[2];
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
void from_cart_to_loc_sphr(double* columnvect_xyzglob, double colatitude, double longitude, double* columnvect_res)
|
||||||
|
{
|
||||||
|
/*IMPORTANT: this subroutine is in the coordinate system NED*/
|
||||||
|
|
||||||
|
double phi = colatitude*DEG2RAD;
|
||||||
|
double lambda = longitude*DEG2RAD;
|
||||||
|
|
||||||
|
double cos_phi = cos(phi);
|
||||||
|
double sin_phi = sin(phi);
|
||||||
|
double cos_lambda = cos(lambda);
|
||||||
|
double sin_lambda = sin(lambda);
|
||||||
|
|
||||||
|
double rowvect_phi_unit[3] = {-sin_phi*cos_lambda, -sin_phi*sin_lambda, cos_phi};
|
||||||
|
double rowvect_lambda_unit[3] = {-sin_lambda, cos_lambda, 0};
|
||||||
|
double rowvect_r_unit[3] = {cos_phi*cos_lambda, cos_phi*sin_lambda, sin_phi};
|
||||||
|
|
||||||
|
columnvect_res[0] = rowvect_phi_unit[0]*columnvect_xyzglob[0]+rowvect_phi_unit[1]*columnvect_xyzglob[1]+rowvect_phi_unit[2]*columnvect_xyzglob[2];
|
||||||
|
columnvect_res[1] = rowvect_lambda_unit[0]*columnvect_xyzglob[0]+rowvect_lambda_unit[1]*columnvect_xyzglob[1]+rowvect_lambda_unit[2]*columnvect_xyzglob[2];
|
||||||
|
columnvect_res[2] = rowvect_r_unit[0]*columnvect_xyzglob[0]+rowvect_r_unit[1]*columnvect_xyzglob[1]+rowvect_r_unit[2]*columnvect_xyzglob[2];
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
void from_loc_sphr_to_loc_sphr(double* columnvect_xyzloc, double colatitude1, double longitude1, double colatitude2, double longitude2, double* columnvect_res)
|
||||||
|
{
|
||||||
|
double columnvect_xyzglob[3];
|
||||||
|
from_loc_sphr_to_cart(columnvect_xyzloc, colatitude1, longitude1, columnvect_xyzglob);
|
||||||
|
from_cart_to_loc_sphr(columnvect_xyzglob, colatitude2, longitude2, columnvect_res);
|
||||||
|
return;
|
||||||
|
|
||||||
|
}
|
||||||
11
dep/magnetic_tesseroids/lib/linalg.h
Executable file
@ -0,0 +1,11 @@
|
|||||||
|
#ifndef _LINALG_H_
|
||||||
|
#define _LINALG_H_
|
||||||
|
|
||||||
|
void conv_vect_cblas(double *vect, double lon1, double lat1, double lon2, double lat2, double *res);
|
||||||
|
void conv_vect_cblas_precalc(double *vect, double cos_a1, double sin_a1, double cos_b1, double sin_b1, double cos_a2, double sin_a2, double cos_b2, double sin_b2, double *res);
|
||||||
|
|
||||||
|
void from_loc_sphr_to_cart(double* columnvect_xyzloc, double colatitude, double longitude, double* columnvect_res);
|
||||||
|
void from_cart_to_loc_sphr(double* columnvect_xyzglob, double colatitude, double longitude, double* columnvect_res);
|
||||||
|
void from_loc_sphr_to_loc_sphr(double* columnvect_xyzloc, double colatitude1, double longitude1, double colatitude2, double longitude2, double* columnvect_res);
|
||||||
|
|
||||||
|
#endif
|
||||||
112
dep/magnetic_tesseroids/lib/logger.cpp
Executable file
@ -0,0 +1,112 @@
|
|||||||
|
/*
|
||||||
|
Functions to set up logging.
|
||||||
|
*/
|
||||||
|
|
||||||
|
|
||||||
|
#include <stdio.h>
|
||||||
|
#include <stdarg.h>
|
||||||
|
#include <time.h>
|
||||||
|
#include "logger.h"
|
||||||
|
|
||||||
|
/* Initialize the logger so that it doesn't print by default */
|
||||||
|
LOGGER logger = {100, 0, 100, NULL};
|
||||||
|
|
||||||
|
|
||||||
|
/* Setup logging to stderr.*/
|
||||||
|
void log_init(int level)
|
||||||
|
{
|
||||||
|
logger.level = level;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Set logging to a file. */
|
||||||
|
void log_tofile(FILE *logfile, int level)
|
||||||
|
{
|
||||||
|
logger.filelogging = 1;
|
||||||
|
logger.logfile = logfile;
|
||||||
|
logger.file_level = level;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Log a message at debug level */
|
||||||
|
void log_debug(const char *fmt, ...)
|
||||||
|
{
|
||||||
|
char msg[10000];
|
||||||
|
va_list args;
|
||||||
|
va_start(args, fmt);
|
||||||
|
vsprintf(msg, fmt, args);
|
||||||
|
va_end(args);
|
||||||
|
|
||||||
|
if(logger.level <= LOG_DEBUG)
|
||||||
|
{
|
||||||
|
fprintf(stderr, "DEBUG: %s\n", msg);
|
||||||
|
}
|
||||||
|
|
||||||
|
if(logger.filelogging && (logger.file_level <= LOG_DEBUG))
|
||||||
|
{
|
||||||
|
fprintf(logger.logfile, "DEBUG: %s\n", msg);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Log a message at info level */
|
||||||
|
void log_info(const char *fmt, ...)
|
||||||
|
{
|
||||||
|
char msg[10000];
|
||||||
|
va_list args;
|
||||||
|
va_start(args, fmt);
|
||||||
|
vsprintf(msg, fmt, args);
|
||||||
|
va_end(args);
|
||||||
|
|
||||||
|
if(logger.level <= LOG_INFO)
|
||||||
|
{
|
||||||
|
fprintf(stderr, "%s\n", msg);
|
||||||
|
}
|
||||||
|
|
||||||
|
if(logger.filelogging && logger.file_level <= LOG_INFO)
|
||||||
|
{
|
||||||
|
fprintf(logger.logfile, "%s\n", msg);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Log a message at warning level */
|
||||||
|
void log_warning(const char *fmt, ...)
|
||||||
|
{
|
||||||
|
char msg[10000];
|
||||||
|
va_list args;
|
||||||
|
va_start(args, fmt);
|
||||||
|
vsprintf(msg, fmt, args);
|
||||||
|
va_end(args);
|
||||||
|
|
||||||
|
if(logger.level <= LOG_WARNING)
|
||||||
|
{
|
||||||
|
fprintf(stderr, "WARNING: %s\n", msg);
|
||||||
|
}
|
||||||
|
|
||||||
|
if(logger.filelogging && logger.file_level <= LOG_WARNING)
|
||||||
|
{
|
||||||
|
fprintf(logger.logfile, "WARNING: %s\n", msg);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Log a message at error level */
|
||||||
|
void log_error(const char *fmt, ...)
|
||||||
|
{
|
||||||
|
char msg[10000];
|
||||||
|
va_list args;
|
||||||
|
va_start(args, fmt);
|
||||||
|
vsprintf(msg, fmt, args);
|
||||||
|
va_end(args);
|
||||||
|
|
||||||
|
if(logger.level <= LOG_ERROR)
|
||||||
|
{
|
||||||
|
fprintf(stderr, "ERROR: %s\n", msg);
|
||||||
|
}
|
||||||
|
|
||||||
|
if(logger.filelogging && logger.file_level <= LOG_ERROR)
|
||||||
|
{
|
||||||
|
fprintf(logger.logfile, "ERROR: %s\n", msg);
|
||||||
|
}
|
||||||
|
}
|
||||||
166
dep/magnetic_tesseroids/lib/logger.h
Executable file
@ -0,0 +1,166 @@
|
|||||||
|
/*
|
||||||
|
Functions to set up logging.
|
||||||
|
|
||||||
|
Examples
|
||||||
|
--------
|
||||||
|
|
||||||
|
Logging to stderr:
|
||||||
|
|
||||||
|
#include "logger.h"
|
||||||
|
|
||||||
|
void my_func(){
|
||||||
|
log_info("From my_func!\n");
|
||||||
|
}
|
||||||
|
|
||||||
|
int main(){
|
||||||
|
// Enable logging to stderr in debug level
|
||||||
|
// will only print messages of level DEBUG or higher
|
||||||
|
log_init(LOG_DEBUG);
|
||||||
|
log_debug("debug line. The code is %d", LOG_DEBUG);
|
||||||
|
log_info("info line. The code is %d", LOG_INFO);
|
||||||
|
log_warning("warning line. The code is %d", LOG_WARNING);
|
||||||
|
log_error("error line. The code is %d", LOG_ERROR);
|
||||||
|
my_func();
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
will print:
|
||||||
|
|
||||||
|
DEBUG: debug line. The code is 0
|
||||||
|
info line. The code is 1
|
||||||
|
WARNING: warning line. The code is 2
|
||||||
|
ERROR: error line. The code is 3
|
||||||
|
From my_func!
|
||||||
|
|
||||||
|
If function log_init() is not called than logging to stderr is disabled and no
|
||||||
|
messages will be printed.
|
||||||
|
|
||||||
|
Logging to a file:
|
||||||
|
|
||||||
|
#include <stdio.h>
|
||||||
|
#include "logger.h"
|
||||||
|
|
||||||
|
void my_func(){
|
||||||
|
log_info("From my_func!\n");
|
||||||
|
log_debug("Should not appear in log file\n");
|
||||||
|
}
|
||||||
|
|
||||||
|
int main(){
|
||||||
|
// Enable logging to file "log.txt" in info level
|
||||||
|
// will not print DEBUG level messages
|
||||||
|
// since log_init was not called, there is no logging to stderr
|
||||||
|
FILE *logfile = fopen("log.txt", "w");
|
||||||
|
log_tofile(logfile, LOG_INFO);
|
||||||
|
log_debug("debug line. The code is %d", LOG_DEBUG);
|
||||||
|
log_info("info line. The code is %d", LOG_INFO);
|
||||||
|
log_warning("warning line. The code is %d", LOG_WARNING);
|
||||||
|
log_error("error line. The code is %d", LOG_ERROR);
|
||||||
|
my_func();
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
File log.txt will look like:
|
||||||
|
|
||||||
|
info line. The code is 1
|
||||||
|
WARNING: warning line. The code is 2
|
||||||
|
ERROR: error line. The code is 3
|
||||||
|
From my_func!
|
||||||
|
|
||||||
|
Note that you can combine loggin to stderr and to a file with different
|
||||||
|
levels in the same program.
|
||||||
|
*/
|
||||||
|
|
||||||
|
#ifndef _TESSEROIDS_LOGGER_H_
|
||||||
|
#define _TESSEROIDS_LOGGER_H_
|
||||||
|
|
||||||
|
/* Needed for definition of FILE */
|
||||||
|
#include <stdio.h>
|
||||||
|
|
||||||
|
|
||||||
|
/** Logging level for debug messages */
|
||||||
|
#define LOG_DEBUG 1
|
||||||
|
/** Logging level for general information */
|
||||||
|
#define LOG_INFO 2
|
||||||
|
/** Logging level for warning messages */
|
||||||
|
#define LOG_WARNING 3
|
||||||
|
/** Logging level for error messages */
|
||||||
|
#define LOG_ERROR 4
|
||||||
|
|
||||||
|
|
||||||
|
/** Keep the information on the global logger */
|
||||||
|
typedef struct logger_struct
|
||||||
|
{
|
||||||
|
int level; /**< level of logging */
|
||||||
|
int filelogging; /**< flag to know wether loggint to a file is enabled */
|
||||||
|
int file_level; /**< logging level for the file */
|
||||||
|
FILE *logfile; /**< file to log to */
|
||||||
|
|
||||||
|
} LOGGER;
|
||||||
|
|
||||||
|
|
||||||
|
/** Global logger struct. Only declare in the main program! */
|
||||||
|
//LOGGER logger;
|
||||||
|
|
||||||
|
|
||||||
|
/** Setup logging to stderr.
|
||||||
|
|
||||||
|
@param level level of logging to be made. Can be one of:
|
||||||
|
- LOG_DEBUG
|
||||||
|
- LOG_INFO
|
||||||
|
- LOG_WARNING
|
||||||
|
- LOG_ERROR
|
||||||
|
*/
|
||||||
|
void log_init(int level);
|
||||||
|
|
||||||
|
|
||||||
|
/** Set logging to a file.
|
||||||
|
|
||||||
|
@param logfile FILE pointer to the already open file to log to.
|
||||||
|
@param level level of logging to be made to the file. Can be one of:
|
||||||
|
- LOG_DEBUG
|
||||||
|
- LOG_INFO
|
||||||
|
- LOG_WARNING
|
||||||
|
- LOG_ERROR
|
||||||
|
*/
|
||||||
|
void log_tofile(FILE *logfile, int level);
|
||||||
|
|
||||||
|
|
||||||
|
/** Log a message at debug level.
|
||||||
|
|
||||||
|
Pass parameters in the same format as printf()
|
||||||
|
|
||||||
|
Prints a newline at the end.
|
||||||
|
*/
|
||||||
|
void log_debug(const char *fmt, ...);
|
||||||
|
|
||||||
|
|
||||||
|
/** Log a message at info level.
|
||||||
|
|
||||||
|
Pass parameters in the same format as printf()
|
||||||
|
|
||||||
|
Does not print "INFO: " in front of the message when logging
|
||||||
|
|
||||||
|
Prints a newline at the end.
|
||||||
|
*/
|
||||||
|
void log_info(const char *fmt, ...);
|
||||||
|
|
||||||
|
|
||||||
|
/** Log a message at warning level.
|
||||||
|
|
||||||
|
Pass parameters in the same format as printf()
|
||||||
|
|
||||||
|
Prints a newline at the end.
|
||||||
|
*/
|
||||||
|
void log_warning(const char *fmt, ...);
|
||||||
|
|
||||||
|
|
||||||
|
/** Log a message at error level.
|
||||||
|
|
||||||
|
Pass parameters in the same format as printf()
|
||||||
|
|
||||||
|
Prints a newline at the end.
|
||||||
|
*/
|
||||||
|
void log_error(const char *fmt, ...);
|
||||||
|
|
||||||
|
|
||||||
|
#endif
|
||||||
678
dep/magnetic_tesseroids/lib/parsers.cpp
Executable file
@ -0,0 +1,678 @@
|
|||||||
|
/*
|
||||||
|
Input and output parsing tools.
|
||||||
|
*/
|
||||||
|
|
||||||
|
|
||||||
|
#include <stdio.h>
|
||||||
|
#include <stdlib.h>
|
||||||
|
#include <string.h>
|
||||||
|
#include <time.h>
|
||||||
|
#include "logger.h"
|
||||||
|
#include "version.h"
|
||||||
|
#include "parsers.h"
|
||||||
|
#include "constants.h"
|
||||||
|
#include "geometry.h"
|
||||||
|
|
||||||
|
#include <math.h>
|
||||||
|
|
||||||
|
|
||||||
|
/* Parse basic command line arguments for programs */
|
||||||
|
int parse_basic_args(int argc, char **argv, const char *progname,
|
||||||
|
BASIC_ARGS *args, void (*print_help)(void))
|
||||||
|
{
|
||||||
|
int bad_args = 0, parsed_args = 0, total_args = 1, i;
|
||||||
|
char *params;
|
||||||
|
|
||||||
|
/* Default values for options */
|
||||||
|
args->verbose = 0;
|
||||||
|
args->logtofile = 0;
|
||||||
|
/* Parse arguments */
|
||||||
|
for(i = 1; i < argc; i++)
|
||||||
|
{
|
||||||
|
if(argv[i][0] == '-')
|
||||||
|
{
|
||||||
|
switch(argv[i][1])
|
||||||
|
{
|
||||||
|
case 'h':
|
||||||
|
if(argv[i][2] != '\0')
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
print_help();
|
||||||
|
return 2;
|
||||||
|
case 'v':
|
||||||
|
if(argv[i][2] != '\0')
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
if(args->verbose)
|
||||||
|
{
|
||||||
|
log_error("repeated option -v");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
args->verbose = 1;
|
||||||
|
break;
|
||||||
|
case 'l':
|
||||||
|
{
|
||||||
|
if(args->logtofile)
|
||||||
|
{
|
||||||
|
log_error("repeated option -l");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
params = &argv[i][2];
|
||||||
|
if(strlen(params) == 0)
|
||||||
|
{
|
||||||
|
log_error("bad input argument -l. Missing filename.");
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
args->logtofile = 1;
|
||||||
|
args->logfname = params;
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
case '-':
|
||||||
|
{
|
||||||
|
params = &argv[i][2];
|
||||||
|
if(strcmp(params, "version"))
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
print_version(progname);
|
||||||
|
return 2;
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
default:
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
if(parsed_args == 0)
|
||||||
|
{
|
||||||
|
args->inputfname = argv[i];
|
||||||
|
parsed_args++;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'. Already given model file %s",
|
||||||
|
argv[i], args->inputfname);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
/* Check if parsing went well */
|
||||||
|
if(parsed_args > total_args)
|
||||||
|
{
|
||||||
|
log_error("%s: too many input arguments. given %d, max %d.",
|
||||||
|
progname, parsed_args, total_args);
|
||||||
|
}
|
||||||
|
if(bad_args > 0)
|
||||||
|
{
|
||||||
|
log_error("%d bad input argument(s)", bad_args);
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
if(parsed_args < total_args)
|
||||||
|
{
|
||||||
|
return 3;
|
||||||
|
}
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
/* Parse command line arguments for tessh* programs */
|
||||||
|
int parse_tessb_args(int argc, char **argv, const char *progname,
|
||||||
|
TESSB_ARGS *args, void (*print_help)(const char *))
|
||||||
|
{
|
||||||
|
int bad_args = 0, parsed_args = 0, total_args = 1, parsed_order = 0,
|
||||||
|
parsed_ratio1 = 0, parsed_ratio2 = 0, parsed_ratio3 = 0, i, nchar, nread;
|
||||||
|
char *params;
|
||||||
|
|
||||||
|
/* Default values for options */
|
||||||
|
args->verbose = 0;
|
||||||
|
args->logtofile = 0;
|
||||||
|
args->lon_order = 2;
|
||||||
|
args->lat_order = 2;
|
||||||
|
args->r_order = 2;
|
||||||
|
args->adaptative = 1;
|
||||||
|
args->ratio1 = 0; /* zero means use the default for the program */
|
||||||
|
args->ratio2 = 0;
|
||||||
|
args->ratio3 = 0;
|
||||||
|
/* Parse arguments */
|
||||||
|
for(i = 1; i < argc; i++)
|
||||||
|
{
|
||||||
|
if(argv[i][0] == '-')
|
||||||
|
{
|
||||||
|
switch(argv[i][1])
|
||||||
|
{
|
||||||
|
case 'h':
|
||||||
|
if(argv[i][2] != '\0')
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
print_help(progname);
|
||||||
|
return 2;
|
||||||
|
case 'v':
|
||||||
|
if(argv[i][2] != '\0')
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
if(args->verbose)
|
||||||
|
{
|
||||||
|
log_error("repeated option -v");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
args->verbose = 1;
|
||||||
|
break;
|
||||||
|
case 'l':
|
||||||
|
{
|
||||||
|
if(args->logtofile)
|
||||||
|
{
|
||||||
|
log_error("repeated option -l");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
params = &argv[i][2];
|
||||||
|
if(strlen(params) == 0)
|
||||||
|
{
|
||||||
|
log_error("bad input argument -l. Missing filename.");
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
args->logtofile = 1;
|
||||||
|
args->logfname = params;
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
case '-':
|
||||||
|
{
|
||||||
|
params = &argv[i][2];
|
||||||
|
if(strcmp(params, "version"))
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
print_version(progname);
|
||||||
|
return 2;
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
case 'a':
|
||||||
|
if(argv[i][2] != '\0')
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
if(!args->adaptative)
|
||||||
|
{
|
||||||
|
log_error("repeated option -a");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
args->adaptative = 0;
|
||||||
|
break;
|
||||||
|
case 'o':
|
||||||
|
{
|
||||||
|
if(parsed_order)
|
||||||
|
{
|
||||||
|
log_error("repeated option -o");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
params = &argv[i][2];
|
||||||
|
nchar = 0;
|
||||||
|
nread = sscanf(params, "%d/%d/%d%n", &(args->lon_order),
|
||||||
|
&(args->lat_order), &(args->r_order), &nchar);
|
||||||
|
if(nread != 3 || *(params + nchar) != '\0')
|
||||||
|
{
|
||||||
|
log_error("bad input argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
parsed_order = 1;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
case 't':
|
||||||
|
{
|
||||||
|
//ELDAR BAYKIEV///////////////////////////////////////////////////////////////////
|
||||||
|
switch(argv[i][2])
|
||||||
|
{
|
||||||
|
case '1':
|
||||||
|
{
|
||||||
|
if(parsed_ratio1)
|
||||||
|
{
|
||||||
|
log_error("repeated option -t1");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
params = &argv[i][3];
|
||||||
|
nchar = 0;
|
||||||
|
nread = sscanf(params, "%lf%n", &(args->ratio1), &nchar);
|
||||||
|
if(nread != 1 || *(params + nchar) != '\0')
|
||||||
|
{
|
||||||
|
log_error("bad input argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
parsed_ratio1 = 1;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
|
||||||
|
case '2':
|
||||||
|
{
|
||||||
|
if(parsed_ratio2)
|
||||||
|
{
|
||||||
|
log_error("repeated option -t2");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
params = &argv[i][3];
|
||||||
|
nchar = 0;
|
||||||
|
nread = sscanf(params, "%lf%n", &(args->ratio2), &nchar);
|
||||||
|
if(nread != 1 || *(params + nchar) != '\0')
|
||||||
|
{
|
||||||
|
log_error("bad input argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
parsed_ratio2 = 1;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
|
||||||
|
case '3':
|
||||||
|
{
|
||||||
|
if(parsed_ratio3)
|
||||||
|
{
|
||||||
|
log_error("repeated option -t3");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
params = &argv[i][3];
|
||||||
|
nchar = 0;
|
||||||
|
nread = sscanf(params, "%lf%n", &(args->ratio3), &nchar);
|
||||||
|
if(nread != 1 || *(params + nchar) != '\0')
|
||||||
|
{
|
||||||
|
log_error("bad input argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
parsed_ratio3 = 1;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
//ELDAR BAYKIEV///////////////////////////////////////////////////////////////////
|
||||||
|
}
|
||||||
|
default:
|
||||||
|
log_error("invalid argument '%s'", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
if(parsed_args == 0)
|
||||||
|
{
|
||||||
|
args->modelfname = argv[i];
|
||||||
|
parsed_args++;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
log_error("invalid argument '%s'. Already given model file %s",
|
||||||
|
argv[i], args->modelfname);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
/* Check if parsing went well */
|
||||||
|
if(bad_args > 0 || parsed_args != total_args)
|
||||||
|
{
|
||||||
|
if(parsed_args < total_args)
|
||||||
|
{
|
||||||
|
log_error("%s: missing input file.",
|
||||||
|
progname, parsed_args, total_args);
|
||||||
|
}
|
||||||
|
if(parsed_args > total_args)
|
||||||
|
{
|
||||||
|
log_error("%s: too many input arguments. given %d, max %d.",
|
||||||
|
progname, parsed_args, total_args);
|
||||||
|
}
|
||||||
|
if(bad_args > 0)
|
||||||
|
{
|
||||||
|
log_error("%d bad input argument(s)", bad_args);
|
||||||
|
}
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
//parse arguments for gradient calculator
|
||||||
|
int parse_gradcalc_args(int argc, char **argv, const char *progname, GRADCALC_ARGS *args, void (*print_help)(const char *))
|
||||||
|
{
|
||||||
|
int bad_args = 0, parsed_args = 0, total_args = 5, i;
|
||||||
|
char *params;
|
||||||
|
|
||||||
|
/* Default values for options */
|
||||||
|
args->verbose = 0;
|
||||||
|
args->logtofile = 0;
|
||||||
|
|
||||||
|
args->gridbx_set = FALSE;
|
||||||
|
args->gridby_set = FALSE;
|
||||||
|
args->gridbz_set = FALSE;
|
||||||
|
args->out_set = -1;
|
||||||
|
|
||||||
|
args->bz_NEU_NED = -1;
|
||||||
|
args->bz_NEU_NED_set = FALSE;
|
||||||
|
|
||||||
|
|
||||||
|
/* Parse arguments */
|
||||||
|
for(i = 1; i < argc; i++)
|
||||||
|
{
|
||||||
|
if(argv[i][0] == '-')
|
||||||
|
{
|
||||||
|
switch(argv[i][1])
|
||||||
|
{
|
||||||
|
case 'h':
|
||||||
|
if(argv[i][2] != '\0')
|
||||||
|
{
|
||||||
|
//log_error("invalid argument '%s'", argv[i]);
|
||||||
|
printf("invalid argument '%s'\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
print_help(progname);
|
||||||
|
return 2;
|
||||||
|
case '-':
|
||||||
|
{
|
||||||
|
params = &argv[i][2];
|
||||||
|
if(strcmp(params, "version"))
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s'\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
print_version(progname);
|
||||||
|
return 2;
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
case 'b':
|
||||||
|
params = &argv[i][2];
|
||||||
|
if(strlen(params) <= 1)
|
||||||
|
{
|
||||||
|
printf("bad input argument -b. Missing component and filename\n");
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
switch(argv[i][2])
|
||||||
|
{
|
||||||
|
case 'x':
|
||||||
|
if(args->gridbx_set)
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s', gridfile bx already set\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
args->gridbx_set = 1;
|
||||||
|
args->gridbx_fn = &argv[i][3];
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
case 'y':
|
||||||
|
if(args->gridby_set)
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s', gridfile by already set\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
args->gridby_set = 1;
|
||||||
|
args->gridby_fn = &argv[i][3];
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
case 'z':
|
||||||
|
if(args->gridbz_set)
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s', gridfile by already set\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
args->gridbz_set = 1;
|
||||||
|
args->gridbz_fn = &argv[i][3];
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
default:
|
||||||
|
printf("invalid argument '%s', wrong component\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
case 'c':
|
||||||
|
params = &argv[i][2];
|
||||||
|
|
||||||
|
if(args->bz_NEU_NED_set)
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s', coordinate system is already set\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
if(strlen(params) > 1)
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s', specify coordinate system in the input grids\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
if(argv[i][2] == '1')
|
||||||
|
{
|
||||||
|
args->bz_NEU_NED_set = 1;
|
||||||
|
args->bz_NEU_NED = 1;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
else if(argv[i][2] == '2')
|
||||||
|
{
|
||||||
|
args->bz_NEU_NED_set = 1;
|
||||||
|
args->bz_NEU_NED = -1;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s', there are only NED (1) and NEU (2, default) coordinate systems\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
break;
|
||||||
|
case 'o':
|
||||||
|
params = &argv[i][2];
|
||||||
|
|
||||||
|
if(args->out_set>=0)
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s', output format is already set\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
|
||||||
|
if(strlen(params) != 1)
|
||||||
|
{
|
||||||
|
printf("invalid argument '%s', specify output format\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
//TODO Add check if it is integer
|
||||||
|
args->out_set = atoi(params);
|
||||||
|
break;
|
||||||
|
default:
|
||||||
|
printf("invalid argument '%s'\n", argv[i]);
|
||||||
|
bad_args++;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
if(parsed_args > total_args)
|
||||||
|
{
|
||||||
|
//log_error("%s: too many input arguments. given %d, max %d.", progname, parsed_args, total_args);
|
||||||
|
}
|
||||||
|
if(bad_args > 0)
|
||||||
|
{
|
||||||
|
//log_error("%d bad input argument(s)", bad_args);
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
if(parsed_args < total_args)
|
||||||
|
{
|
||||||
|
return 3;
|
||||||
|
}
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
/* Strip trailing spaces and newlines from the end of a string */
|
||||||
|
void strstrip(char *str)
|
||||||
|
{
|
||||||
|
int i;
|
||||||
|
for(i = strlen(str) - 1; i >= 0; i--)
|
||||||
|
{
|
||||||
|
if(str[i] != ' ' && str[i] != '\n' && str[i] != '\r' && str[i] != '\0')
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
str[i + 1] = '\0';
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Read a single tesseroid from a string */
|
||||||
|
int gets_mag_tess(const char *str, MAG_TESSEROID *tess)
|
||||||
|
{
|
||||||
|
double w, e, s, n, top, bot, dens, suscept, Bx, By, Bz, Rx, Ry, Rz;
|
||||||
|
int nread, nchars;
|
||||||
|
|
||||||
|
nread = sscanf(str, "%lf %lf %lf %lf %lf %lf %lf %lf %lf %lf %lf%n", &w, &e, &s,
|
||||||
|
&n, &top, &bot, &dens, &suscept, &Bx, &By, &Bz, &nchars);
|
||||||
|
if(nread != 11 || str[nchars] != '\0')
|
||||||
|
{
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
tess->w = w;
|
||||||
|
tess->e = e;
|
||||||
|
tess->s = s;
|
||||||
|
tess->n = n;
|
||||||
|
tess->r1 = MEAN_EARTH_RADIUS + bot;
|
||||||
|
tess->r2 = MEAN_EARTH_RADIUS + top;
|
||||||
|
tess->density = dens;
|
||||||
|
tess->suscept = suscept;
|
||||||
|
tess->Bx = Bx;
|
||||||
|
tess->By = By;
|
||||||
|
tess->Bz = Bz;
|
||||||
|
|
||||||
|
tess->cos_a1 = cos(PI/2.0-DEG2RAD*(w+e)*0.5);
|
||||||
|
tess->sin_a1 = sin(PI/2.0-DEG2RAD*(w+e)*0.5);
|
||||||
|
tess->cos_b1 = cos(DEG2RAD*(s+n)*0.5);
|
||||||
|
tess->sin_b1 = sin(DEG2RAD*(s+n)*0.5);
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
|
||||||
|
//ELDAR BAYKIEV////////////////////////////////
|
||||||
|
MAG_TESSEROID * read_mag_tess_model(FILE *modelfile, int *size)
|
||||||
|
{
|
||||||
|
MAG_TESSEROID *model, *tmp;
|
||||||
|
int buffsize = 300, line, badinput = 0, error_exit = 0;
|
||||||
|
char sbuff[10000];
|
||||||
|
|
||||||
|
/* Start with a single buffer allocation and expand later if necessary */
|
||||||
|
model = (MAG_TESSEROID *)malloc(buffsize*sizeof(MAG_TESSEROID));
|
||||||
|
if(model == NULL)
|
||||||
|
{
|
||||||
|
log_error("problem allocating initial memory to load tesseroid model.");
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
*size = 0;
|
||||||
|
for(line = 1; !feof(modelfile); line++)
|
||||||
|
{
|
||||||
|
if(fgets(sbuff, 10000, modelfile) == NULL)
|
||||||
|
{
|
||||||
|
if(ferror(modelfile))
|
||||||
|
{
|
||||||
|
log_error("problem encountered reading line %d.", line);
|
||||||
|
error_exit = 1;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
/* Check for comments and blank lines */
|
||||||
|
if(sbuff[0] == '#' || sbuff[0] == '\r' || sbuff[0] == '\n')
|
||||||
|
{
|
||||||
|
continue;
|
||||||
|
}
|
||||||
|
if(*size == buffsize)
|
||||||
|
{
|
||||||
|
buffsize += buffsize;
|
||||||
|
tmp = (MAG_TESSEROID *)realloc(model, buffsize*sizeof(MAG_TESSEROID));
|
||||||
|
if(tmp == NULL)
|
||||||
|
{
|
||||||
|
/* Need to free because realloc leaves unchanged in case of
|
||||||
|
error */
|
||||||
|
free(model);
|
||||||
|
log_error("problem expanding memory for tesseroid model.\nModel is too big.");
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
model = tmp;
|
||||||
|
}
|
||||||
|
/* Remove any trailing spaces or newlines */
|
||||||
|
strstrip(sbuff);
|
||||||
|
if(gets_mag_tess(sbuff, &model[*size]))
|
||||||
|
{
|
||||||
|
log_warning("bad/invalid tesseroid at line %d.", line);
|
||||||
|
badinput = 1;
|
||||||
|
continue;
|
||||||
|
}
|
||||||
|
(*size)++;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
if(badinput || error_exit)
|
||||||
|
{
|
||||||
|
free(model);
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
/* Adjust the size of the model */
|
||||||
|
if(*size != 0)
|
||||||
|
{
|
||||||
|
tmp = (MAG_TESSEROID *)realloc(model, (*size)*sizeof(MAG_TESSEROID));
|
||||||
|
if(tmp == NULL)
|
||||||
|
{
|
||||||
|
/* Need to free because realloc leaves unchanged in case of
|
||||||
|
error */
|
||||||
|
free(model);
|
||||||
|
log_error("problem freeing excess memory for tesseroid model.");
|
||||||
|
return NULL;
|
||||||
|
}
|
||||||
|
model = tmp;
|
||||||
|
}
|
||||||
|
return model;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Read a single rectangular prism from a string */
|
||||||
75
dep/magnetic_tesseroids/lib/parsers.h
Executable file
@ -0,0 +1,75 @@
|
|||||||
|
/*
|
||||||
|
Input and output parsing tools.
|
||||||
|
*/
|
||||||
|
|
||||||
|
|
||||||
|
#ifndef _MAG_TESSEROIDS_PARSERS_H_
|
||||||
|
#define _MAG_TESSEROIDS_PARSERS_H_
|
||||||
|
|
||||||
|
/* Needed for definition of MAG_TESSEROID and PRISM */
|
||||||
|
#include "geometry.h"
|
||||||
|
/* Need for the definition of FILE */
|
||||||
|
#include <stdio.h>
|
||||||
|
|
||||||
|
/** Store basic input arguments and option flags */
|
||||||
|
typedef struct basic_args
|
||||||
|
{
|
||||||
|
char *inputfname; /**< name of the input file */
|
||||||
|
int verbose; /**< flag to indicate if verbose printing is enabled */
|
||||||
|
int logtofile; /**< flag to indicate if logging to a file is enabled */
|
||||||
|
char *logfname; /**< name of the log file */
|
||||||
|
} BASIC_ARGS;
|
||||||
|
|
||||||
|
typedef struct tessh_args
|
||||||
|
{
|
||||||
|
int lon_order; /**< glq order in longitude integration */
|
||||||
|
int lat_order; /**< glq order in latitude integration */
|
||||||
|
int r_order; /**< glq order in radial integration */
|
||||||
|
char *modelfname; /**< name of the file with the tesseroid model */
|
||||||
|
int verbose; /**< flag to indicate if verbose printing is enabled */
|
||||||
|
int logtofile; /**< flag to indicate if logging to a file is enabled */
|
||||||
|
char *logfname; /**< name of the log file */
|
||||||
|
int adaptative; /**< flat to indicate wether to use the adaptative size
|
||||||
|
of tesseroid algorithm */
|
||||||
|
double ratio1; /**< distance-size ratio used for recusive division */
|
||||||
|
double ratio2; /**< distance-size ratio used for recusive division */
|
||||||
|
double ratio3; /**< distance-size ratio used for recusive division */
|
||||||
|
} TESSB_ARGS;
|
||||||
|
|
||||||
|
|
||||||
|
typedef struct gradcalc_args
|
||||||
|
{
|
||||||
|
int gridbx_set;
|
||||||
|
int gridby_set;
|
||||||
|
int gridbz_set;
|
||||||
|
|
||||||
|
char* gridbx_fn;
|
||||||
|
char* gridby_fn;
|
||||||
|
char* gridbz_fn;
|
||||||
|
|
||||||
|
int out_set;
|
||||||
|
|
||||||
|
|
||||||
|
int bz_NEU_NED;
|
||||||
|
int bz_NEU_NED_set;
|
||||||
|
|
||||||
|
int verbose; /**< flag to indicate if verbose printing is enabled */
|
||||||
|
int logtofile; /**< flag to indicate if logging to a file is enabled */
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
} GRADCALC_ARGS;
|
||||||
|
|
||||||
|
int parse_basic_args(int argc, char **argv, const char *progname, BASIC_ARGS *args, void (*print_help)(void));
|
||||||
|
int parse_tessb_args(int argc, char **argv, const char *progname, TESSB_ARGS *args, void (*print_help)(const char *));
|
||||||
|
|
||||||
|
int parse_gradcalc_args(int argc, char **argv, const char *progname, GRADCALC_ARGS *args, void (*print_help)(const char *));
|
||||||
|
|
||||||
|
void strstrip(char *str);
|
||||||
|
|
||||||
|
int gets_mag_tess(const char *str, MAG_TESSEROID *tess);
|
||||||
|
MAG_TESSEROID * read_mag_tess_model(FILE *modelfile, int *size);
|
||||||
|
|
||||||
|
|
||||||
|
#endif
|
||||||
385
dep/magnetic_tesseroids/lib/tessb_main.cpp
Executable file
@ -0,0 +1,385 @@
|
|||||||
|
/*
|
||||||
|
Generic main function for the tessb* programs.
|
||||||
|
*/
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
#include <stdio.h>
|
||||||
|
#include <stdlib.h>
|
||||||
|
#include <string.h>
|
||||||
|
#include <time.h>
|
||||||
|
#include "logger.h"
|
||||||
|
#include "version.h"
|
||||||
|
#include "grav_tess.h"
|
||||||
|
#include "glq.h"
|
||||||
|
#include "constants.h"
|
||||||
|
#include "geometry.h"
|
||||||
|
#include "parsers.h"
|
||||||
|
#include "tessb_main.h"
|
||||||
|
#include "linalg.h"
|
||||||
|
|
||||||
|
#include <math.h>
|
||||||
|
|
||||||
|
/* Print the help message for tessh* programs */
|
||||||
|
void print_tessb_help(const char *progname)
|
||||||
|
{
|
||||||
|
printf("MAGNETIC MAG_TESSEROIDS\n");
|
||||||
|
printf("Usage: %s MODELFILE [OPTIONS]\n\n", progname);
|
||||||
|
if(strcmp(progname + 4, "pot") == 0)
|
||||||
|
{
|
||||||
|
printf("Calculate the potential due to a tesseroid model on\n");
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
printf("Calculate the %s component due to a tesseroid model on\n",
|
||||||
|
progname + 4);
|
||||||
|
}
|
||||||
|
printf("TODO\n");
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* Run the main for a generic tessh* program */
|
||||||
|
int run_tessb_main(int argc, char **argv, const char *progname,
|
||||||
|
double (*field)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ),
|
||||||
|
double ratio1, double ratio2, double ratio3)
|
||||||
|
{
|
||||||
|
TESSB_ARGS args;
|
||||||
|
GLQ *glq_lon, *glq_lat, *glq_r;
|
||||||
|
MAG_TESSEROID *model;
|
||||||
|
|
||||||
|
int modelsize, rc, line, points = 0, error_exit = 0, bad_input = 0;
|
||||||
|
char buff[10000];
|
||||||
|
|
||||||
|
double lon, lat, height, res;
|
||||||
|
|
||||||
|
/*variables for precalculation of trigonometrical functions for tesseroid centers*/
|
||||||
|
double cos_a2, sin_a2, cos_b2, sin_b2;
|
||||||
|
|
||||||
|
/*variables for precalculation of trigonometrical functions for grid points*/
|
||||||
|
double lon_prev, lat_prev;
|
||||||
|
double cos_a2_prev, sin_a2_prev, cos_b2_prev, sin_b2_prev;
|
||||||
|
|
||||||
|
FILE *logfile = NULL, *modelfile = NULL;
|
||||||
|
time_t rawtime;
|
||||||
|
clock_t tstart;
|
||||||
|
struct tm * timeinfo;
|
||||||
|
|
||||||
|
double (*field1)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ);
|
||||||
|
double (*field2)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ);
|
||||||
|
double (*field3)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ);
|
||||||
|
void (*field_triple)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ, double*);
|
||||||
|
double ggt_1, ggt_2, ggt_3;
|
||||||
|
double gtt_v[3];
|
||||||
|
int n_tesseroid;
|
||||||
|
|
||||||
|
|
||||||
|
log_init(LOG_INFO);
|
||||||
|
|
||||||
|
rc = parse_tessb_args(argc, argv, progname, &args, &print_tessb_help);
|
||||||
|
if(rc == 2)
|
||||||
|
{
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
if(rc == 1)
|
||||||
|
{
|
||||||
|
log_warning("Terminating due to bad input");
|
||||||
|
log_warning("Try '%s -h' for instructions", progname);
|
||||||
|
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
|
||||||
|
/* Set the appropriate logging level and log to file if necessary */
|
||||||
|
if(!args.verbose)
|
||||||
|
{
|
||||||
|
log_init(LOG_WARNING);
|
||||||
|
}
|
||||||
|
if(args.logtofile)
|
||||||
|
{
|
||||||
|
logfile = fopen(args.logfname, "w");
|
||||||
|
if(logfile == NULL)
|
||||||
|
{
|
||||||
|
log_error("unable to create log file %s", args.logfname);
|
||||||
|
log_warning("Terminating due to bad input");
|
||||||
|
log_warning("Try '%s -h' for instructions", progname);
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
log_tofile(logfile, LOG_DEBUG);
|
||||||
|
}
|
||||||
|
|
||||||
|
/* Check if a custom distance-size ratio is given */
|
||||||
|
if(args.ratio1 != 0)
|
||||||
|
{
|
||||||
|
ratio1 = args.ratio1;
|
||||||
|
}
|
||||||
|
if(args.ratio2 != 0)
|
||||||
|
{
|
||||||
|
ratio2 = args.ratio2;
|
||||||
|
}
|
||||||
|
if(args.ratio3 != 0)
|
||||||
|
{
|
||||||
|
ratio3 = args.ratio3;
|
||||||
|
}
|
||||||
|
|
||||||
|
/* Print standard verbose */
|
||||||
|
log_info("%s (Tesseroids project) %s", progname, tesseroids_version);
|
||||||
|
time(&rawtime);
|
||||||
|
timeinfo = localtime(&rawtime);
|
||||||
|
log_info("(local time) %s", asctime(timeinfo));
|
||||||
|
log_info("Use recursive division of tesseroids: %s",
|
||||||
|
args.adaptative ? "True" : "False");
|
||||||
|
log_info("Distance-size ratio1 for recusive division: %g", ratio1);
|
||||||
|
log_info("Distance-size ratio2 for recusive division: %g", ratio2);
|
||||||
|
log_info("Distance-size ratio3 for recusive division: %g", ratio3);
|
||||||
|
|
||||||
|
/* Make the necessary GLQ structures */
|
||||||
|
log_info("Using GLQ orders: %d lon / %d lat / %d r", args.lon_order,
|
||||||
|
args.lat_order, args.r_order);
|
||||||
|
glq_lon = glq_new(args.lon_order, -1, 1);
|
||||||
|
glq_lat = glq_new(args.lat_order, -1, 1);
|
||||||
|
glq_r = glq_new(args.r_order, -1, 1);
|
||||||
|
if(glq_lon == NULL || glq_lat == NULL || glq_r == NULL)
|
||||||
|
{
|
||||||
|
log_error("failed to create required GLQ structures");
|
||||||
|
log_warning("Terminating due to bad input");
|
||||||
|
log_warning("Try '%s -h' for instructions", progname);
|
||||||
|
if(args.logtofile)
|
||||||
|
fclose(logfile);
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
|
||||||
|
/* Read the tesseroid model file */
|
||||||
|
log_info("Reading magnetic tesseroid model from file %s", args.modelfname);
|
||||||
|
modelfile = fopen(args.modelfname, "r");
|
||||||
|
if(modelfile == NULL)
|
||||||
|
{
|
||||||
|
log_error("failed to open model file %s", args.modelfname);
|
||||||
|
log_warning("Terminating due to bad input");
|
||||||
|
log_warning("Try '%s -h' for instructions", progname);
|
||||||
|
if(args.logtofile)
|
||||||
|
fclose(logfile);
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
model = read_mag_tess_model(modelfile, &modelsize);
|
||||||
|
fclose(modelfile);
|
||||||
|
if(modelsize == 0)
|
||||||
|
{
|
||||||
|
log_error("tesseroid file %s is empty", args.modelfname);
|
||||||
|
log_warning("Terminating due to bad input");
|
||||||
|
log_warning("Try '%s -h' for instructions", progname);
|
||||||
|
if(args.logtofile)
|
||||||
|
fclose(logfile);
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
if(model == NULL)
|
||||||
|
{
|
||||||
|
log_error("failed to read model from file %s", args.modelfname);
|
||||||
|
log_warning("Terminating due to bad input");
|
||||||
|
log_warning("Try '%s -h' for instructions", progname);
|
||||||
|
if(args.logtofile)
|
||||||
|
fclose(logfile);
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
log_info("Total of %d tesseroid(s) read", modelsize);
|
||||||
|
|
||||||
|
/* Print a header on the output with provenance information */
|
||||||
|
if(strcmp(progname + 4, "pot") == 0)
|
||||||
|
{
|
||||||
|
printf("# Potential calculated with %s %s:\n", progname,
|
||||||
|
tesseroids_version);
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
printf("# %s component calculated with %s %s:\n", progname+4, progname,
|
||||||
|
tesseroids_version);
|
||||||
|
}
|
||||||
|
printf("# local time: %s", asctime(timeinfo));
|
||||||
|
printf("# model file: %s (%d tesseroids)\n", args.modelfname, modelsize);
|
||||||
|
printf("# GLQ order: %d lon / %d lat / %d r\n", args.lon_order,
|
||||||
|
args.lat_order, args.r_order);
|
||||||
|
printf("# Use recursive division of tesseroids: %s\n",
|
||||||
|
args.adaptative ? "True" : "False");
|
||||||
|
printf("# Distance-size ratio1 for recusive division: %g\n", ratio1);
|
||||||
|
printf("# Distance-size ratio2 for recusive division: %g\n", ratio2);
|
||||||
|
printf("# Distance-size ratio3 for recusive division: %g\n", ratio3);
|
||||||
|
|
||||||
|
/////////////ELDAR BAYKIEV//////////////
|
||||||
|
/* Assign pointers to functions that calculate gravity gradient tensor components */
|
||||||
|
if (!strcmp("tessbx", progname))
|
||||||
|
{
|
||||||
|
field1 = &tess_gxx;
|
||||||
|
field2 = &tess_gxy;
|
||||||
|
field3 = &tess_gxz;
|
||||||
|
|
||||||
|
field_triple = &tess_gxx_gxy_gxz;
|
||||||
|
}
|
||||||
|
|
||||||
|
if (!strcmp("tessby", progname))
|
||||||
|
{
|
||||||
|
field1 = &tess_gxy;
|
||||||
|
field2 = &tess_gyy;
|
||||||
|
field3 = &tess_gyz;
|
||||||
|
|
||||||
|
field_triple = &tess_gxy_gyy_gyz;
|
||||||
|
}
|
||||||
|
|
||||||
|
if (!strcmp("tessbz", progname))
|
||||||
|
{
|
||||||
|
field1 = &tess_gxz;
|
||||||
|
field2 = &tess_gyz;
|
||||||
|
field3 = &tess_gzz;
|
||||||
|
|
||||||
|
field_triple = &tess_gxz_gyz_gzz;
|
||||||
|
}
|
||||||
|
/////////////ELDAR BAYKIEV//////////////
|
||||||
|
|
||||||
|
/* Read each computation point from stdin and calculate */
|
||||||
|
log_info("Calculating (this may take a while)...");
|
||||||
|
tstart = clock();
|
||||||
|
|
||||||
|
///////////////
|
||||||
|
lon_prev = 0;
|
||||||
|
lat_prev = 0;
|
||||||
|
|
||||||
|
for(line = 1; !feof(stdin); line++)
|
||||||
|
{
|
||||||
|
if(fgets(buff, 10000, stdin) == NULL)
|
||||||
|
{
|
||||||
|
if(ferror(stdin))
|
||||||
|
{
|
||||||
|
log_error("problem encountered reading line %d", line);
|
||||||
|
error_exit = 1;
|
||||||
|
break;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
/* Check for comments and blank lines */
|
||||||
|
if(buff[0] == '#' || buff[0] == '\r' || buff[0] == '\n')
|
||||||
|
{
|
||||||
|
printf("%s", buff);
|
||||||
|
continue;
|
||||||
|
}
|
||||||
|
if(sscanf(buff, "%lf %lf %lf", &lon, &lat, &height) != 3)
|
||||||
|
{
|
||||||
|
log_warning("bad/invalid computation point at line %d", line);
|
||||||
|
log_warning("skipping this line and continuing");
|
||||||
|
bad_input++;
|
||||||
|
continue;
|
||||||
|
}
|
||||||
|
/* Need to remove \n and \r from end of buff first to print the
|
||||||
|
result in the end */
|
||||||
|
strstrip(buff);
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
/////////////ELDAR BAYKIEV//////////////
|
||||||
|
res = 0;
|
||||||
|
if(args.adaptative)
|
||||||
|
{
|
||||||
|
for(n_tesseroid = 0; n_tesseroid < modelsize; n_tesseroid++)
|
||||||
|
{
|
||||||
|
gtt_v[0] = 0;
|
||||||
|
gtt_v[1] = 0;
|
||||||
|
gtt_v[2] = 0;
|
||||||
|
double B_to_H = model[n_tesseroid].suscept/(M_0);//IMPORTANT
|
||||||
|
double M_vect[3] = {model[n_tesseroid].Bx * B_to_H, model[n_tesseroid].By * B_to_H, model[n_tesseroid].Bz * B_to_H};
|
||||||
|
double M_vect_p[3] = {0, 0, 0};
|
||||||
|
|
||||||
|
conv_vect_cblas(M_vect, (model[n_tesseroid].w + model[n_tesseroid].e)*0.5, (model[n_tesseroid].s + model[n_tesseroid].n)*0.5, lon, lat, M_vect_p);
|
||||||
|
|
||||||
|
ggt_1 = calc_tess_model_adapt(&model[n_tesseroid], 1, lon, lat, height + MEAN_EARTH_RADIUS, glq_lon, glq_lat, glq_r, field1, ratio1);
|
||||||
|
ggt_2 = calc_tess_model_adapt(&model[n_tesseroid], 1, lon, lat, height + MEAN_EARTH_RADIUS, glq_lon, glq_lat, glq_r, field2, ratio2);
|
||||||
|
ggt_3 = calc_tess_model_adapt(&model[n_tesseroid], 1, lon, lat, height + MEAN_EARTH_RADIUS, glq_lon, glq_lat, glq_r, field3, ratio3);
|
||||||
|
|
||||||
|
res = res + M_0*EOTVOS2SI*(ggt_1 * M_vect_p[0] + ggt_2 * M_vect_p[1] + ggt_3 * M_vect_p[2]) /(G*model[n_tesseroid].density*4*PI);
|
||||||
|
|
||||||
|
//printf("res %g\n", res);
|
||||||
|
}
|
||||||
|
|
||||||
|
/////////////////////////////////////////////////////////////////////////////////////////////////////////
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
//precalculate trigonometrical functions
|
||||||
|
|
||||||
|
if(lon == lon_prev)
|
||||||
|
{
|
||||||
|
cos_b2 = cos_b2_prev;
|
||||||
|
sin_b2 = sin_b2_prev;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
cos_b2 = cos(DEG2RAD*lon);
|
||||||
|
sin_b2 = sin(DEG2RAD*lon);
|
||||||
|
}
|
||||||
|
|
||||||
|
if(lat == lat_prev)
|
||||||
|
{
|
||||||
|
cos_a2 = cos_a2_prev;
|
||||||
|
sin_a2 = sin_a2_prev;
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
cos_a2 = cos(PI/2.0-DEG2RAD*lat);
|
||||||
|
sin_a2 = sin(PI/2.0-DEG2RAD*lat);
|
||||||
|
}
|
||||||
|
/////////////////////////////////////////////////////////////////////////////////////////////////////////
|
||||||
|
for(n_tesseroid = 0; n_tesseroid < modelsize; n_tesseroid++)
|
||||||
|
{
|
||||||
|
gtt_v[0] = 0;
|
||||||
|
gtt_v[1] = 0;
|
||||||
|
gtt_v[2] = 0;
|
||||||
|
//ELDAR: TODO: PRECALCULATE SIC COSINE TABLES
|
||||||
|
double B_to_H = model[n_tesseroid].suscept/(M_0);//IMPORTANT
|
||||||
|
double M_vect[3] = {model[n_tesseroid].Bx * B_to_H, model[n_tesseroid].By * B_to_H, model[n_tesseroid].Bz * B_to_H};
|
||||||
|
double M_vect_p[3] = {0, 0, 0};
|
||||||
|
|
||||||
|
//conv_vect_cblas(M_vect, (model[n_tesseroid].w + model[n_tesseroid].e)*0.5, (model[n_tesseroid].s + model[n_tesseroid].n)*0.5, lon, lat, M_vect_p);
|
||||||
|
conv_vect_cblas_precalc(M_vect, model[n_tesseroid].cos_a1, model[n_tesseroid].sin_a1, model[n_tesseroid].cos_b1, model[n_tesseroid].sin_b1, cos_a2, sin_a2, cos_b2, sin_b2, M_vect_p);
|
||||||
|
|
||||||
|
calc_tess_model_triple(&model[n_tesseroid], 1, lon, lat, height + MEAN_EARTH_RADIUS, glq_lon, glq_lat, glq_r, field_triple, gtt_v);
|
||||||
|
|
||||||
|
res = res + M_0*EOTVOS2SI*(gtt_v[0] * M_vect_p[0] + gtt_v[1] * M_vect_p[1] + gtt_v[2] * M_vect_p[2]) /(G*model[n_tesseroid].density*4*PI);
|
||||||
|
|
||||||
|
}
|
||||||
|
|
||||||
|
lon_prev = lon;
|
||||||
|
lat_prev = lat;
|
||||||
|
|
||||||
|
cos_a2_prev = cos_a2;
|
||||||
|
sin_a2_prev = sin_a2;
|
||||||
|
|
||||||
|
cos_b2_prev = cos_b2;
|
||||||
|
sin_b2_prev = sin_b2;
|
||||||
|
/////////////////////////////////////////////////////////////////////////////////////////////////////////
|
||||||
|
}
|
||||||
|
printf("%s %.15g\n", buff, res);
|
||||||
|
points++;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
if(bad_input)
|
||||||
|
{
|
||||||
|
log_warning("Encountered %d bad computation points which were skipped",
|
||||||
|
bad_input);
|
||||||
|
}
|
||||||
|
if(error_exit)
|
||||||
|
{
|
||||||
|
log_warning("Terminating due to error in input");
|
||||||
|
log_warning("Try '%s -h' for instructions", progname);
|
||||||
|
}
|
||||||
|
else
|
||||||
|
{
|
||||||
|
log_info("Calculated on %d points in %.5g seconds", points,
|
||||||
|
(double)(clock() - tstart)/CLOCKS_PER_SEC);
|
||||||
|
}
|
||||||
|
/* Clean up */
|
||||||
|
free(model);
|
||||||
|
glq_free(glq_lon);
|
||||||
|
glq_free(glq_lat);
|
||||||
|
glq_free(glq_r);
|
||||||
|
log_info("Done");
|
||||||
|
if(args.logtofile)
|
||||||
|
fclose(logfile);
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
14
dep/magnetic_tesseroids/lib/tessb_main.h
Executable file
@ -0,0 +1,14 @@
|
|||||||
|
/*
|
||||||
|
Generic main function for the tessb* programs.
|
||||||
|
*/
|
||||||
|
|
||||||
|
#ifndef _MAG_TESSEROIDS_TESSH_MAIN_H_
|
||||||
|
#define _MAG_TESSEROIDS_TESSH_MAIN_H_
|
||||||
|
|
||||||
|
#include "glq.h"
|
||||||
|
#include "geometry.h"
|
||||||
|
|
||||||
|
void print_tessb_help(const char *progname);
|
||||||
|
int run_tessb_main(int argc, char **argv, const char *progname, double (*field)(MAG_TESSEROID, double, double, double, GLQ, GLQ, GLQ), double ratio1, double ratio2, double ratio3);
|
||||||
|
|
||||||
|
#endif
|
||||||
17
dep/magnetic_tesseroids/lib/version.cpp
Executable file
@ -0,0 +1,17 @@
|
|||||||
|
/*
|
||||||
|
Hold the version number of the project.
|
||||||
|
*/
|
||||||
|
|
||||||
|
#include <stdio.h>
|
||||||
|
#include "version.h"
|
||||||
|
|
||||||
|
|
||||||
|
/* Print version and license information */
|
||||||
|
void print_version(const char *progname)
|
||||||
|
{
|
||||||
|
printf("%s (Magnetic Tesseroids) %s\n\n", progname, tesseroids_version);
|
||||||
|
printf("Copyright (C) 2015, Eldar Baykiev.\n");
|
||||||
|
printf("This program is based on the code of Leonardo Uieda.\n");
|
||||||
|
printf("<http://tesseroids.readthedocs.org/>\n");
|
||||||
|
printf("Developed by Eldar Baykiev.\n");
|
||||||
|
}
|
||||||
18
dep/magnetic_tesseroids/lib/version.h
Executable file
@ -0,0 +1,18 @@
|
|||||||
|
/*
|
||||||
|
Hold the version number of the project.
|
||||||
|
*/
|
||||||
|
|
||||||
|
#ifndef _TESSEROIDS_VERSION_H_
|
||||||
|
#define _TESSEROIDS_VERSION_H_
|
||||||
|
|
||||||
|
/** Current project version number */
|
||||||
|
const char tesseroids_version[] = "1.1";
|
||||||
|
|
||||||
|
|
||||||
|
/** Print version and license information
|
||||||
|
|
||||||
|
@param progname name of the program
|
||||||
|
*/
|
||||||
|
void print_version(const char *progname);
|
||||||
|
|
||||||
|
#endif
|
||||||
3
dep/magnetic_tesseroids/stow-config.sh
Executable file
@ -0,0 +1,3 @@
|
|||||||
|
#!/bin/bash
|
||||||
|
|
||||||
|
sudo stow --dir=/opt/stow --target=/usr/local -S magtess
|
||||||
36
dep/magnetic_tesseroids/toolkits/CMakeLists.txt
Normal file
@ -0,0 +1,36 @@
|
|||||||
|
# 设置编译选项
|
||||||
|
set(CMAKE_C_FLAGS "${CMAKE_C_FLAGS} -O3")
|
||||||
|
set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} -O3")
|
||||||
|
|
||||||
|
# 设置可执行文件的输出地址
|
||||||
|
set(EXECUTABLE_OUTPUT_PATH ${PROJECT_BINARY_DIR}/bin/toolkits)
|
||||||
|
|
||||||
|
# 下面设置tools的编译命令 首先设置一个宏
|
||||||
|
macro(add_tools name)
|
||||||
|
# 添加可执行程序名称
|
||||||
|
add_executable(${name} ${name}.cpp)
|
||||||
|
# 设置安装后的动态库调用地址
|
||||||
|
set_target_properties(${name} PROPERTIES INSTALL_RPATH /usr/local/lib)
|
||||||
|
# 链接动态库
|
||||||
|
target_link_libraries(${name} PUBLIC magtess)
|
||||||
|
# 将可执行程序安装到bin
|
||||||
|
install(TARGETS ${name} RUNTIME DESTINATION sbin)
|
||||||
|
endmacro()
|
||||||
|
|
||||||
|
# 添加tools
|
||||||
|
add_tools(tessbx)
|
||||||
|
add_tools(tessby)
|
||||||
|
add_tools(tessbz)
|
||||||
|
add_tools(tessutil_combine_grids)
|
||||||
|
add_tools(tessutil_gradient_calculator)
|
||||||
|
|
||||||
|
if(${CMAKE_HOST_SYSTEM_NAME} STREQUAL "Linux" OR ${CMAKE_HOST_SYSTEM_NAME} STREQUAL "Darwin")
|
||||||
|
# 添加可执行程序名称
|
||||||
|
add_executable(tessutil_magnetize_model tessutil_magnetize_model.c)
|
||||||
|
# 设置安装后的动态库调用地址
|
||||||
|
set_target_properties(tessutil_magnetize_model PROPERTIES INSTALL_RPATH /usr/local/lib)
|
||||||
|
# 链接动态库
|
||||||
|
target_link_libraries(tessutil_magnetize_model PUBLIC magtess)
|
||||||
|
# 将可执行程序安装到bin
|
||||||
|
install(TARGETS tessutil_magnetize_model RUNTIME DESTINATION sbin)
|
||||||
|
endif()
|
||||||
11
dep/magnetic_tesseroids/toolkits/tessbx.cpp
Executable file
@ -0,0 +1,11 @@
|
|||||||
|
#include "../lib/constants.h"
|
||||||
|
#include "../lib/grav_tess.h"
|
||||||
|
#include "../lib/tessb_main.h"
|
||||||
|
|
||||||
|
|
||||||
|
/** Main tessbx*/
|
||||||
|
int main(int argc, char **argv)
|
||||||
|
{
|
||||||
|
return run_tessb_main(argc, argv, "tessbx", 0, TESSEROID_GXX_SIZE_RATIO, TESSEROID_GXY_SIZE_RATIO, TESSEROID_GXZ_SIZE_RATIO);
|
||||||
|
|
||||||
|
}
|
||||||
11
dep/magnetic_tesseroids/toolkits/tessby.cpp
Executable file
@ -0,0 +1,11 @@
|
|||||||
|
#include "../lib/constants.h"
|
||||||
|
#include "../lib/grav_tess.h"
|
||||||
|
#include "../lib/tessb_main.h"
|
||||||
|
|
||||||
|
|
||||||
|
/** Main tessby*/
|
||||||
|
int main(int argc, char **argv)
|
||||||
|
{
|
||||||
|
return run_tessb_main(argc, argv, "tessby", 0, TESSEROID_GXX_SIZE_RATIO, TESSEROID_GXY_SIZE_RATIO, TESSEROID_GXZ_SIZE_RATIO);
|
||||||
|
|
||||||
|
}
|
||||||
11
dep/magnetic_tesseroids/toolkits/tessbz.cpp
Executable file
@ -0,0 +1,11 @@
|
|||||||
|
#include "../lib/constants.h"
|
||||||
|
#include "../lib/grav_tess.h"
|
||||||
|
#include "../lib/tessb_main.h"
|
||||||
|
|
||||||
|
|
||||||
|
/** Main tessbz*/
|
||||||
|
int main(int argc, char **argv)
|
||||||
|
{
|
||||||
|
return run_tessb_main(argc, argv, "tessbz", 0, TESSEROID_GXX_SIZE_RATIO, TESSEROID_GXY_SIZE_RATIO, TESSEROID_GXZ_SIZE_RATIO);
|
||||||
|
|
||||||
|
}
|
||||||
102
dep/magnetic_tesseroids/toolkits/tessutil_combine_grids.cpp
Executable file
@ -0,0 +1,102 @@
|
|||||||
|
#include <stdio.h>
|
||||||
|
#include <stdlib.h>
|
||||||
|
#include <math.h>
|
||||||
|
#include <fstream>
|
||||||
|
#include <iostream> // Added by Zhang Yi on 2021-08-26
|
||||||
|
#include <string>
|
||||||
|
|
||||||
|
#define MAX_GRID_POINTS 16000
|
||||||
|
|
||||||
|
#define GRID_FORMAT "%lf %lf %f %lf"
|
||||||
|
|
||||||
|
#if defined(_MSC_VER) /* Added for windows by Yi Zhang on 2021-08-26*/
|
||||||
|
#include <BaseTsd.h>
|
||||||
|
typedef SSIZE_T ssize_t;
|
||||||
|
#endif
|
||||||
|
|
||||||
|
void printresult_withalt(double* longitudes, double* latitudes, float* altitudes, double* values, int n_values)
|
||||||
|
{
|
||||||
|
|
||||||
|
|
||||||
|
for (int h = 0; h < n_values; h++)
|
||||||
|
printf( "%lf %lf %f %lf\n", longitudes[h], latitudes[h], altitudes[h], values[h]);
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
void printresult(double* longitudes, double* latitudes, double* values, int n_values)
|
||||||
|
{
|
||||||
|
|
||||||
|
|
||||||
|
for (int h = 0; h < n_values; h++)
|
||||||
|
printf( "%lf %lf %lf\n", longitudes[h], latitudes[h], values[h]);
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
int main(int argc, char**argv)
|
||||||
|
{
|
||||||
|
int n_files = (argc-1)/2;
|
||||||
|
|
||||||
|
double lons[MAX_GRID_POINTS];
|
||||||
|
double lats[MAX_GRID_POINTS];
|
||||||
|
float alts[MAX_GRID_POINTS];
|
||||||
|
|
||||||
|
double vals[MAX_GRID_POINTS];
|
||||||
|
|
||||||
|
double factor = 1;
|
||||||
|
|
||||||
|
//char * line = NULL;
|
||||||
|
std::string line;
|
||||||
|
size_t len = 0;
|
||||||
|
ssize_t read;
|
||||||
|
|
||||||
|
int n_lines = 0;
|
||||||
|
|
||||||
|
for (int i = 0; i < n_files; i++)
|
||||||
|
{
|
||||||
|
//FILE * fp = fopen(argv[1+2*i], "r");
|
||||||
|
std::ifstream fp(argv[1+2*i], std::ios::in);
|
||||||
|
sscanf(argv[1+2*i+1], "%lf", &factor);
|
||||||
|
|
||||||
|
//if (fp == NULL)
|
||||||
|
if (!fp)
|
||||||
|
{
|
||||||
|
printf("ERROR: Can not open file with grid values.\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
n_lines = 0;
|
||||||
|
//while ((read = getline(&line, &len, fp )) != -1)
|
||||||
|
while (std::getline(fp, line))
|
||||||
|
{
|
||||||
|
|
||||||
|
//if ((line[0] != '#') && (strlen(line) > 2))
|
||||||
|
if ((line[0] != '#') && (line.length() > 2))
|
||||||
|
{
|
||||||
|
n_lines++;
|
||||||
|
if (n_lines>MAX_GRID_POINTS)
|
||||||
|
{
|
||||||
|
printf("ERROR: Too many grid points (> %d) in the input. Recompile program with a bigger value of MAX_GRID_POINTS.\n", n_lines);
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
double value;
|
||||||
|
|
||||||
|
sscanf(line.c_str(), GRID_FORMAT, &lons[n_lines-1], &lats[n_lines-1], &alts[n_lines-1], &value);
|
||||||
|
vals[n_lines-1] = vals[n_lines-1] + value*factor;
|
||||||
|
|
||||||
|
}
|
||||||
|
}
|
||||||
|
//fclose(fp);
|
||||||
|
fp.close();
|
||||||
|
}
|
||||||
|
|
||||||
|
int no_alt = 0;
|
||||||
|
|
||||||
|
if (no_alt)
|
||||||
|
printresult(lons, lats, vals, n_lines);
|
||||||
|
else
|
||||||
|
printresult_withalt(lons, lats, alts, vals, n_lines);
|
||||||
|
|
||||||
|
}
|
||||||
366
dep/magnetic_tesseroids/toolkits/tessutil_gradient_calculator.cpp
Executable file
@ -0,0 +1,366 @@
|
|||||||
|
#include <stdio.h>
|
||||||
|
#include <stdlib.h>
|
||||||
|
#include <math.h>
|
||||||
|
#include <fstream>
|
||||||
|
#include <iostream> // Added by Zhang Yi on 2021-08-26
|
||||||
|
#include <string>
|
||||||
|
|
||||||
|
#include "../lib/constants.h"
|
||||||
|
#include "../lib/parsers.h"
|
||||||
|
#include "../lib/linalg.h"
|
||||||
|
|
||||||
|
#define MAX_GRID_POINTS 50000
|
||||||
|
|
||||||
|
#define GRID_FORMAT "%lf %lf %f %lf"
|
||||||
|
|
||||||
|
#if defined(_MSC_VER) /* Added for windows by Yi Zhang on 2021-08-26*/
|
||||||
|
#include <BaseTsd.h>
|
||||||
|
typedef SSIZE_T ssize_t;
|
||||||
|
#endif
|
||||||
|
|
||||||
|
|
||||||
|
// TODO conversion of input/output units nT/km pT/km nT/m pT/m
|
||||||
|
|
||||||
|
|
||||||
|
void printcomp(double* longitudes, double* latitudes, double* values, int n_values)
|
||||||
|
{
|
||||||
|
|
||||||
|
|
||||||
|
for (int h = 0; h < n_values; h++)
|
||||||
|
printf("%lf %lf %lf\n", longitudes[h], latitudes[h], values[h]);
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
void printall(double* longitudes, double* latitudes, int n_values, double* values1, double* values2, double* values3, double* values4, double* values5, double* values6, double* values7)
|
||||||
|
{
|
||||||
|
|
||||||
|
|
||||||
|
for (int h = 0; h < n_values; h++)
|
||||||
|
printf("%lf %lf %lf %lf %lf %lf %lf %lf %lf\n", longitudes[h], latitudes[h], values1[h], values2[h], values3[h], values4[h], values5[h], values6[h], values7[h]);
|
||||||
|
|
||||||
|
return;
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
void print_gradcalc_help(const char *progname)
|
||||||
|
{
|
||||||
|
printf("MAGNETIC TESSEROIDS: Gradient Calculator\n");
|
||||||
|
printf("Usage: %s [OPTIONS]\n\n", progname);
|
||||||
|
printf("Options:\n");
|
||||||
|
printf("\t-h\t\t\t Help\n\n");
|
||||||
|
|
||||||
|
printf("\t-bx[GRID FILENAME]\t Grid filename with Bx component\n");
|
||||||
|
printf("\t-by[GRID FILENAME]\t Grid filename with By component\n");
|
||||||
|
printf("\t-bz[GRID FILENAME]\t Grid filename with Bz component\n");
|
||||||
|
printf("\tNOTE:\tall grids must be in format LON LAT ALT B*,\n\t\tstart from West-South corner and longitudes must increment first.\n\t\tLON, LAT in [deg], B* in [nT] and ALT in [m].\n\n");
|
||||||
|
|
||||||
|
printf("\t-c[COORD SYSTEM]\t Coordinate system in input grids. 1 - North-East-Down, 2 - North-East-Up\n");
|
||||||
|
printf("\t-o[COMPONENT]\t\t If 0, then output format is LON LAT BXX BYX BZX BXY BYY BZY BZZ, if 1-7, then \n");
|
||||||
|
printf("\tonly corresponding component would be printed with format LON LAT B**.\n");
|
||||||
|
printf("\tNOTE: output is always in North-East-Down coordinate system.\n\t\tLON, LAT in [deg], B** in [nT/km].\n");
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
//printf("TODO\n");
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
int main(int argc, char**argv)
|
||||||
|
{
|
||||||
|
const char * progname = "tessutil_gradient_calculator";
|
||||||
|
GRADCALC_ARGS args;
|
||||||
|
int rc;
|
||||||
|
|
||||||
|
|
||||||
|
//log_init(LOG_INFO);
|
||||||
|
|
||||||
|
rc = parse_gradcalc_args(argc, argv, progname, &args, &print_gradcalc_help);
|
||||||
|
if(rc == 2) //help
|
||||||
|
{
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
|
if(rc == 1)
|
||||||
|
{
|
||||||
|
printf("Terminating due to bad input\n");
|
||||||
|
printf("Try '%s -h' for instructions\n", progname);
|
||||||
|
|
||||||
|
return 1;
|
||||||
|
}
|
||||||
|
|
||||||
|
if ((args.gridbx_set == 0) || (args.gridbx_set == 0) || (args.gridbx_set == 0))
|
||||||
|
{
|
||||||
|
printf("no input grids\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
if (args.bz_NEU_NED == 1)
|
||||||
|
printf("#Coordinate system in input grids: North-East-Down\n");
|
||||||
|
else
|
||||||
|
printf("#Coordinate system in input grids: North-East-Up\n");
|
||||||
|
printf("#Coordinate system in output grid: North-East-Down\n");
|
||||||
|
|
||||||
|
|
||||||
|
double lons[MAX_GRID_POINTS];
|
||||||
|
double lats[MAX_GRID_POINTS];
|
||||||
|
float alts[MAX_GRID_POINTS];
|
||||||
|
|
||||||
|
double bx[MAX_GRID_POINTS];
|
||||||
|
double by[MAX_GRID_POINTS];
|
||||||
|
double bz[MAX_GRID_POINTS];
|
||||||
|
//read file with bx
|
||||||
|
|
||||||
|
//char * line = NULL;
|
||||||
|
std::string line;
|
||||||
|
size_t len = 0;
|
||||||
|
ssize_t read;
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
//FILE * bxfp = fopen(args.gridbx_fn, "r");
|
||||||
|
//if (bxfp == NULL)
|
||||||
|
std::ifstream bxfp(args.gridbx_fn, std::ios::in); // Updated by Yi Zhang on 2021-08-26
|
||||||
|
if (!bxfp)
|
||||||
|
{
|
||||||
|
printf("ERROR: Can not open file with Bx values.\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
int n_lines = 0;
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
//while ((read = getline(&line, &len, bxfp )) != -1)
|
||||||
|
while (getline(bxfp, line))
|
||||||
|
{
|
||||||
|
|
||||||
|
//if ((line[0] != '#') && (strlen(line) > 2))
|
||||||
|
if ((line[0] != '#') && (line.length() > 2))
|
||||||
|
{
|
||||||
|
n_lines++;
|
||||||
|
if (n_lines>MAX_GRID_POINTS)
|
||||||
|
{
|
||||||
|
printf("ERROR: Too many grid points (> %d) in the input. Recompile program with a bigger value of MAX_GRID_POINTS.\n", n_lines);
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
sscanf(line.c_str(), GRID_FORMAT, &lons[n_lines-1], &lats[n_lines-1], &alts[n_lines-1], &bx[n_lines-1]);
|
||||||
|
|
||||||
|
}
|
||||||
|
}
|
||||||
|
//fclose(bxfp);
|
||||||
|
bxfp.close();
|
||||||
|
|
||||||
|
|
||||||
|
/*number of grid points*/
|
||||||
|
printf("#Number of grid points: %d\n", n_lines);
|
||||||
|
|
||||||
|
/*grid spacing*/
|
||||||
|
|
||||||
|
double lon_min = lons[0];
|
||||||
|
double lon_max = lons[n_lines-1];
|
||||||
|
|
||||||
|
double lon_step = lons[1]-lons[0];
|
||||||
|
if ((lon_step == 0) || (lon_step <= 0) || (lon_max < lon_min))
|
||||||
|
{
|
||||||
|
printf("ERROR: Wrong grid format. Longitudes must increment first. Use the format of tessgrd.\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
int lon_n = 1;
|
||||||
|
while ((lats[lon_n] == lats[0]) && (lon_n < n_lines))
|
||||||
|
{
|
||||||
|
lon_n++;
|
||||||
|
}
|
||||||
|
double lat_step = lats[lon_n]-lats[0];
|
||||||
|
|
||||||
|
int lat_n = n_lines / lon_n;
|
||||||
|
|
||||||
|
double lat_min = lats[0];
|
||||||
|
double lat_max = lats[n_lines-1];
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
if ((lat_step <= 0) || (lat_step <= 0) || (lat_max < lat_min))
|
||||||
|
{
|
||||||
|
printf("ERROR: Wrong grid format. Latitudes must increment. Use the format of tessgrd.\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
printf("#Longitudinal step: %lf, latitudinal step: %lf \n", lon_step, lat_step);
|
||||||
|
printf("#Longitudinal points: %d, latitudinal points: %d \n", lon_n, lat_n);
|
||||||
|
printf("#Edges: W %lf, E %lf, S %lf, N %lf \n", lon_min, lon_max, lat_min, lat_max);
|
||||||
|
|
||||||
|
/* read other grids */
|
||||||
|
// By
|
||||||
|
//FILE * byfp = fopen(args.gridby_fn, "r");
|
||||||
|
//if (byfp == NULL)
|
||||||
|
|
||||||
|
std::ifstream byfp(args.gridby_fn, std::ios::in);
|
||||||
|
if (!byfp)
|
||||||
|
{
|
||||||
|
printf("ERROR: Can not open file with Bx values.\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
int n_lines2 = 0;
|
||||||
|
//while ((read = getline(&line, &len, byfp )) != -1)
|
||||||
|
while (getline(byfp, line))
|
||||||
|
{
|
||||||
|
|
||||||
|
//if ((line[0] != '#') && (strlen(line) > 2))
|
||||||
|
if ((line[0] != '#') && (line.length() > 2))
|
||||||
|
{
|
||||||
|
n_lines2++;
|
||||||
|
//printf("%s", line);
|
||||||
|
double dummy1, dummy2;
|
||||||
|
float dummy3;
|
||||||
|
sscanf(line.c_str(), GRID_FORMAT , &dummy1, &dummy2, &dummy3, &by[n_lines2-1]);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
//fclose(byfp);
|
||||||
|
byfp.close();
|
||||||
|
|
||||||
|
if (n_lines2 != n_lines)
|
||||||
|
{
|
||||||
|
printf("ERROR: Grid points of Bx and By do not coincide.\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
// Bz
|
||||||
|
//FILE * bzfp = fopen(args.gridbz_fn, "r");
|
||||||
|
//if (bzfp == NULL)
|
||||||
|
std::ifstream bzfp(args.gridbz_fn, std::ios::in);
|
||||||
|
if (!bzfp)
|
||||||
|
{
|
||||||
|
printf("ERROR: Can not open file with Bx values.\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
n_lines2 = 0;
|
||||||
|
//while ((read = getline(&line, &len, bzfp )) != -1)
|
||||||
|
while (getline(bzfp, line))
|
||||||
|
{
|
||||||
|
if ((line[0] != '#') && (line.length() > 2))
|
||||||
|
{
|
||||||
|
n_lines2++;
|
||||||
|
//printf("%s", line);
|
||||||
|
double dummy1, dummy2;
|
||||||
|
float dummy3;
|
||||||
|
double bz_curr;
|
||||||
|
sscanf(line.c_str(), GRID_FORMAT, &dummy1, &dummy2, &dummy3,&bz_curr);
|
||||||
|
|
||||||
|
bz[n_lines2-1] = args.bz_NEU_NED* bz_curr; //COORDINATE SYSTEM NEU or NED
|
||||||
|
}
|
||||||
|
}
|
||||||
|
//fclose(byfp);
|
||||||
|
byfp.close();
|
||||||
|
|
||||||
|
if (n_lines2 != n_lines)
|
||||||
|
{
|
||||||
|
printf("ERROR: Grid points of Bx and Bz do not coincide.\n");
|
||||||
|
exit(EXIT_FAILURE);
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
/* calculate gradients */
|
||||||
|
|
||||||
|
|
||||||
|
double bxx[MAX_GRID_POINTS];
|
||||||
|
double byx[MAX_GRID_POINTS];
|
||||||
|
double bzx[MAX_GRID_POINTS];
|
||||||
|
|
||||||
|
double bxy[MAX_GRID_POINTS];
|
||||||
|
double byy[MAX_GRID_POINTS];
|
||||||
|
double bzy[MAX_GRID_POINTS];
|
||||||
|
|
||||||
|
double bzz[MAX_GRID_POINTS];
|
||||||
|
|
||||||
|
int cent_ind, west_ind, east_ind, south_ind, north_ind;
|
||||||
|
|
||||||
|
for (int i = 1; i<lon_n-1; i++)
|
||||||
|
{
|
||||||
|
for (int j = 1; j<lat_n-1; j++)
|
||||||
|
{
|
||||||
|
cent_ind = (j) *lon_n + (i);
|
||||||
|
west_ind = (j) *lon_n + (i-1);
|
||||||
|
east_ind = (j) *lon_n + (i+1);
|
||||||
|
south_ind = (j-1) *lon_n + (i);
|
||||||
|
north_ind = (j+1) *lon_n + (i);
|
||||||
|
|
||||||
|
//double vect_cent[3] = {bx[cent_ind], by[cent_ind], bz[cent_ind]};
|
||||||
|
double vect_west[3] = {bx[west_ind], by[west_ind], bz[west_ind]};
|
||||||
|
double vect_east[3] = {bx[east_ind], by[east_ind], bz[east_ind]};
|
||||||
|
double vect_south[3] = {bx[south_ind], by[south_ind], bz[south_ind]};
|
||||||
|
double vect_north[3] = {bx[north_ind], by[north_ind], bz[north_ind]};
|
||||||
|
|
||||||
|
double vect_c_west[3];
|
||||||
|
double vect_c_east[3];
|
||||||
|
double vect_c_south[3];
|
||||||
|
double vect_c_north[3];
|
||||||
|
|
||||||
|
from_loc_sphr_to_loc_sphr(vect_west, lats[west_ind], lons[west_ind], lats[cent_ind], lons[cent_ind], vect_c_west);
|
||||||
|
from_loc_sphr_to_loc_sphr(vect_east, lats[east_ind], lons[east_ind], lats[cent_ind], lons[cent_ind], vect_c_east);
|
||||||
|
from_loc_sphr_to_loc_sphr(vect_south, lats[south_ind], lons[south_ind], lats[cent_ind], lons[cent_ind], vect_c_south);
|
||||||
|
from_loc_sphr_to_loc_sphr(vect_north, lats[north_ind], lons[north_ind], lats[cent_ind], lons[cent_ind], vect_c_north);
|
||||||
|
|
||||||
|
double cent_ang_sn = acos(sin(lats[south_ind]*DEG2RAD)*sin(lats[north_ind]*DEG2RAD) + cos(lats[south_ind]*DEG2RAD)*cos(lats[north_ind]*DEG2RAD)*cos(abs(lons[south_ind]-lons[north_ind])*DEG2RAD));
|
||||||
|
double cent_ang_we = acos(sin(lats[west_ind]*DEG2RAD)*sin(lats[east_ind]*DEG2RAD) + cos(lats[west_ind]*DEG2RAD)*cos(lats[east_ind]*DEG2RAD)*cos(abs(lons[west_ind]-lons[east_ind])*DEG2RAD));
|
||||||
|
|
||||||
|
double dist_sn = (MEAN_EARTH_RADIUS + alts[cent_ind])*cent_ang_sn;
|
||||||
|
double dist_we = (MEAN_EARTH_RADIUS + alts[cent_ind])*cent_ang_we;
|
||||||
|
|
||||||
|
bxx[cent_ind] = -(vect_c_south[0]-vect_c_north[0])/dist_sn*1000.0;
|
||||||
|
byx[cent_ind] = -(vect_c_south[1]-vect_c_north[1])/dist_sn*1000.0;
|
||||||
|
bzx[cent_ind] = -(vect_c_south[2]-vect_c_north[2])/dist_sn*1000.0;
|
||||||
|
|
||||||
|
bxy[cent_ind] = -(vect_c_west[0]-vect_c_east[0])/dist_we*1000.0;
|
||||||
|
byy[cent_ind] = -(vect_c_west[1]-vect_c_east[1])/dist_we*1000.0;
|
||||||
|
bzy[cent_ind] = -(vect_c_west[2]-vect_c_east[2])/dist_we*1000.0;
|
||||||
|
|
||||||
|
bzz[cent_ind] = -bxx[cent_ind] - byy[cent_ind];
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
switch(args.out_set)
|
||||||
|
{
|
||||||
|
case 1:
|
||||||
|
printf("#Component Bxx\n");
|
||||||
|
printcomp(lons, lats, bxx, n_lines);
|
||||||
|
break;
|
||||||
|
case 2:
|
||||||
|
printf("#Component Byx\n");
|
||||||
|
printcomp(lons, lats, byx, n_lines);
|
||||||
|
break;
|
||||||
|
case 3:
|
||||||
|
printf("#Component Bzx\n");
|
||||||
|
printcomp(lons, lats, bzx, n_lines);
|
||||||
|
break;
|
||||||
|
case 4:
|
||||||
|
printf("#Component Bxy\n");
|
||||||
|
printcomp(lons, lats, bxy, n_lines);
|
||||||
|
break;
|
||||||
|
case 5:
|
||||||
|
printf("#Component Byy\n");
|
||||||
|
printcomp(lons, lats, byy, n_lines);
|
||||||
|
break;
|
||||||
|
case 6:
|
||||||
|
printf("#Component Bzy\n");
|
||||||
|
printcomp(lons, lats, bzy, n_lines);
|
||||||
|
break;
|
||||||
|
case 7:
|
||||||
|
printf("#Component Bzz\n");
|
||||||
|
printcomp(lons, lats, bzz, n_lines);
|
||||||
|
break;
|
||||||
|
default:
|
||||||
|
printf("#All components: Bxx, Byx, Bzx, Bxy, Byy, Bzy, Bzz\n");
|
||||||
|
printall(lons, lats, n_lines, bxx, byx, bzx, bxy, byy, bzy, bzz);
|
||||||
|
break;
|
||||||
|
|
||||||
|
}
|
||||||
|
|
||||||
|
}
|
||||||
1318
dep/magnetic_tesseroids/toolkits/tessutil_magnetize_model.c
Executable file
14
dep/netcdfcxx_legacy/CMakeLists.txt
Normal file
@ -0,0 +1,14 @@
|
|||||||
|
cmake_minimum_required(VERSION 3.15.2)
|
||||||
|
# 设置工程名称和语言
|
||||||
|
project(netCDF_CXX_LEGACY VERSION 1.0)
|
||||||
|
# 添加配置配件编写的函数
|
||||||
|
include(CMakePackageConfigHelpers)
|
||||||
|
|
||||||
|
message(STATUS "Platform: " ${CMAKE_HOST_SYSTEM_NAME})
|
||||||
|
# CMake默认的安装路径 Windows下为C:/Program\ Files/${Project_Name} Linux/Unix下为/usr/local
|
||||||
|
message(STATUS "Install prefix: " ${CMAKE_INSTALL_PREFIX})
|
||||||
|
# CMake默认的变异类型为空
|
||||||
|
message(STATUS "Build type: " ${CMAKE_BUILD_TYPE})
|
||||||
|
|
||||||
|
# 添加源文件地址
|
||||||
|
add_subdirectory(src/)
|
||||||
21
dep/netcdfcxx_legacy/LICENSE
Normal file
@ -0,0 +1,21 @@
|
|||||||
|
MIT License
|
||||||
|
|
||||||
|
Copyright (c) 2021 yizhangss
|
||||||
|
|
||||||
|
Permission is hereby granted, free of charge, to any person obtaining a copy
|
||||||
|
of this software and associated documentation files (the "Software"), to deal
|
||||||
|
in the Software without restriction, including without limitation the rights
|
||||||
|
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
||||||
|
copies of the Software, and to permit persons to whom the Software is
|
||||||
|
furnished to do so, subject to the following conditions:
|
||||||
|
|
||||||
|
The above copyright notice and this permission notice shall be included in all
|
||||||
|
copies or substantial portions of the Software.
|
||||||
|
|
||||||
|
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
||||||
|
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
||||||
|
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
||||||
|
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
||||||
|
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
||||||
|
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
||||||
|
SOFTWARE.
|
||||||
36
dep/netcdfcxx_legacy/README.en.md
Normal file
@ -0,0 +1,36 @@
|
|||||||
|
# netcdfcxx_win
|
||||||
|
|
||||||
|
#### Description
|
||||||
|
一个windows下netcdf c++接口的编译项目,注意使用的是4.2版本的老接口。
|
||||||
|
|
||||||
|
#### Software Architecture
|
||||||
|
Software architecture description
|
||||||
|
|
||||||
|
#### Installation
|
||||||
|
|
||||||
|
1. xxxx
|
||||||
|
2. xxxx
|
||||||
|
3. xxxx
|
||||||
|
|
||||||
|
#### Instructions
|
||||||
|
|
||||||
|
1. xxxx
|
||||||
|
2. xxxx
|
||||||
|
3. xxxx
|
||||||
|
|
||||||
|
#### Contribution
|
||||||
|
|
||||||
|
1. Fork the repository
|
||||||
|
2. Create Feat_xxx branch
|
||||||
|
3. Commit your code
|
||||||
|
4. Create Pull Request
|
||||||
|
|
||||||
|
|
||||||
|
#### Gitee Feature
|
||||||
|
|
||||||
|
1. You can use Readme\_XXX.md to support different languages, such as Readme\_en.md, Readme\_zh.md
|
||||||
|
2. Gitee blog [blog.gitee.com](https://blog.gitee.com)
|
||||||
|
3. Explore open source project [https://gitee.com/explore](https://gitee.com/explore)
|
||||||
|
4. The most valuable open source project [GVP](https://gitee.com/gvp)
|
||||||
|
5. The manual of Gitee [https://gitee.com/help](https://gitee.com/help)
|
||||||
|
6. The most popular members [https://gitee.com/gitee-stars/](https://gitee.com/gitee-stars/)
|
||||||
37
dep/netcdfcxx_legacy/README.md
Normal file
@ -0,0 +1,37 @@
|
|||||||
|
# netcdfcxx_legacy
|
||||||
|
|
||||||
|
#### 介绍
|
||||||
|
一个windows下netcdf c++接口的编译项目,注意使用的是4.2版本的老接口。
|
||||||
|
|
||||||
|
#### 软件架构
|
||||||
|
软件架构说明
|
||||||
|
|
||||||
|
|
||||||
|
#### 安装教程
|
||||||
|
|
||||||
|
1. xxxx
|
||||||
|
2. xxxx
|
||||||
|
3. xxxx
|
||||||
|
|
||||||
|
#### 使用说明
|
||||||
|
|
||||||
|
1. xxxx
|
||||||
|
2. xxxx
|
||||||
|
3. xxxx
|
||||||
|
|
||||||
|
#### 参与贡献
|
||||||
|
|
||||||
|
1. Fork 本仓库
|
||||||
|
2. 新建 Feat_xxx 分支
|
||||||
|
3. 提交代码
|
||||||
|
4. 新建 Pull Request
|
||||||
|
|
||||||
|
|
||||||
|
#### 特技
|
||||||
|
|
||||||
|
1. 使用 Readme\_XXX.md 来支持不同的语言,例如 Readme\_en.md, Readme\_zh.md
|
||||||
|
2. Gitee 官方博客 [blog.gitee.com](https://blog.gitee.com)
|
||||||
|
3. 你可以 [https://gitee.com/explore](https://gitee.com/explore) 这个地址来了解 Gitee 上的优秀开源项目
|
||||||
|
4. [GVP](https://gitee.com/gvp) 全称是 Gitee 最有价值开源项目,是综合评定出的优秀开源项目
|
||||||
|
5. Gitee 官方提供的使用手册 [https://gitee.com/help](https://gitee.com/help)
|
||||||
|
6. Gitee 封面人物是一档用来展示 Gitee 会员风采的栏目 [https://gitee.com/gitee-stars/](https://gitee.com/gitee-stars/)
|
||||||
17
dep/netcdfcxx_legacy/config.sh
Executable file
@ -0,0 +1,17 @@
|
|||||||
|
#!/bin/bash
|
||||||
|
|
||||||
|
cmd=${1}
|
||||||
|
package=netcdfcxx_legacy
|
||||||
|
address=/opt/stow
|
||||||
|
targetdir=/usr/local
|
||||||
|
|
||||||
|
if [[ ${cmd} == "configure" && ! -d "build/" ]]; then
|
||||||
|
mkdir build && cd build && cmake .. -DCMAKE_INSTALL_PREFIX=${address}/${package} -DCMAKE_BUILD_TYPE=Release
|
||||||
|
elif [[ ${cmd} == "configure" ]]; then
|
||||||
|
cd build && rm -rf * && cmake .. -DCMAKE_INSTALL_PREFIX=${address}/${package} -DCMAKE_BUILD_TYPE=Release
|
||||||
|
elif [[ ${cmd} == "build" ]]; then
|
||||||
|
cd build && make
|
||||||
|
elif [[ ${cmd} == "install" ]]; then
|
||||||
|
cd build && sudo make install
|
||||||
|
sudo stow --dir=${address} --target=${targetdir} ${package}
|
||||||
|
fi
|
||||||
18
dep/netcdfcxx_legacy/netCDF_CXX_LEGACYConfig.cmake.in
Normal file
@ -0,0 +1,18 @@
|
|||||||
|
@PACKAGE_INIT@
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_Version "@PROJECT_VERSION@")
|
||||||
|
set_and_check(@PROJECT_NAME@_INSTALL_PREFIX "${PACKAGE_PREFIX_DIR}")
|
||||||
|
set_and_check(@PROJECT_NAME@_INC_DIR "${PACKAGE_PREFIX_DIR}/include")
|
||||||
|
set_and_check(@PROJECT_NAME@_INCULDE_DIR "${PACKAGE_PREFIX_DIR}/include")
|
||||||
|
set_and_check(@PROJECT_NAME@_LIB_DIR "${PACKAGE_PREFIX_DIR}/lib")
|
||||||
|
set_and_check(@PROJECT_NAME@_LIBRARY_DIR "${PACKAGE_PREFIX_DIR}/lib")
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_LIB netcdfcxx_legacy)
|
||||||
|
set(@PROJECT_NAME@_LIBRARY netcdfcxx_legacy)
|
||||||
|
set(@PROJECT_NAME@_FOUND 1)
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_OPENMP @LibLCG_OPENMP@)
|
||||||
|
set(@PROJECT_NAME@_EIGEN @LibLCG_EIGEN@)
|
||||||
|
|
||||||
|
# include target information
|
||||||
|
include("${CMAKE_CURRENT_LIST_DIR}/@PROJECT_NAME@Targets.cmake")
|
||||||
79
dep/netcdfcxx_legacy/src/CMakeLists.txt
Normal file
@ -0,0 +1,79 @@
|
|||||||
|
# 设定源文件文件夹
|
||||||
|
aux_source_directory(lib/ NETCDF_SRC)
|
||||||
|
|
||||||
|
# 寻找netCDF库地址
|
||||||
|
find_package(netCDF REQUIRED)
|
||||||
|
if(netCDF_FOUND)
|
||||||
|
include_directories(${netCDF_INCLUDE_DIR})
|
||||||
|
endif()
|
||||||
|
|
||||||
|
# 设置编译选项
|
||||||
|
set(CMAKE_C_FLAGS "${CMAKE_C_FLAGS} -O3")
|
||||||
|
set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} -O3")
|
||||||
|
# 设置库文件的输出地址
|
||||||
|
set(LIBRARY_OUTPUT_PATH ${PROJECT_BINARY_DIR}/lib)
|
||||||
|
|
||||||
|
# 以下部分为库的编译
|
||||||
|
# 注意目标名必须唯一 所以不能直接生成相同名称的动态库与静态库
|
||||||
|
# 注意此处不必为目标名称添加lib前缀和相应后缀,cmake会自行添加
|
||||||
|
add_library(netcdfcxx_legacy SHARED ${NETCDF_SRC})
|
||||||
|
# 首先添加静态库的生成命令
|
||||||
|
add_library(netcdfcxx_legacy_static STATIC ${NETCDF_SRC})
|
||||||
|
# 设置静态库的输出名称从而获得与动态库名称相同的静态库
|
||||||
|
set_target_properties(netcdfcxx_legacy_static PROPERTIES OUTPUT_NAME "netcdfcxx_legacy")
|
||||||
|
# 设置输出目标属性以同时输出动态库与静态库
|
||||||
|
set_target_properties(netcdfcxx_legacy PROPERTIES CLEAN_DIRECT_OUTPUT 1)
|
||||||
|
set_target_properties(netcdfcxx_legacy_static PROPERTIES CLEAN_DIRECT_OUTPUT 1)
|
||||||
|
# 设置动态库的版本号
|
||||||
|
set_target_properties(netcdfcxx_legacy PROPERTIES VERSION ${PROJECT_VERSION} SOVERSION ${PROJECT_VERSION_MAJOR}.${PROJECT_VERSION_MINOR})
|
||||||
|
|
||||||
|
find_library(NETCDF_LIBRARY netcdf ${netCDF_LIB_DIR})
|
||||||
|
target_link_libraries(netcdfcxx_legacy PUBLIC ${NETCDF_LIBRARY})
|
||||||
|
target_link_libraries(netcdfcxx_legacy_static ${NETCDF_LIBRARY})
|
||||||
|
|
||||||
|
set(CONFIG_FILE_PATH lib/cmake/${PROJECT_NAME})
|
||||||
|
|
||||||
|
configure_package_config_file(${PROJECT_SOURCE_DIR}/${PROJECT_NAME}Config.cmake.in
|
||||||
|
${CMAKE_BINARY_DIR}/${PROJECT_NAME}Config.cmake
|
||||||
|
INSTALL_DESTINATION ${CONFIG_FILE_PATH}
|
||||||
|
NO_CHECK_REQUIRED_COMPONENTS_MACRO)
|
||||||
|
|
||||||
|
write_basic_package_version_file(${CMAKE_BINARY_DIR}/${PROJECT_NAME}ConfigVersion.cmake
|
||||||
|
VERSION ${PROJECT_VERSION}
|
||||||
|
COMPATIBILITY SameMajorVersion)
|
||||||
|
|
||||||
|
# 库的安装命令
|
||||||
|
install(TARGETS netcdfcxx_legacy netcdfcxx_legacy_static
|
||||||
|
EXPORT ${PROJECT_NAME}Targets
|
||||||
|
LIBRARY DESTINATION lib
|
||||||
|
ARCHIVE DESTINATION lib)
|
||||||
|
install(EXPORT ${PROJECT_NAME}Targets
|
||||||
|
DESTINATION ${CONFIG_FILE_PATH})
|
||||||
|
install(FILES
|
||||||
|
${CMAKE_BINARY_DIR}/${PROJECT_NAME}Config.cmake
|
||||||
|
${CMAKE_BINARY_DIR}/${PROJECT_NAME}ConfigVersion.cmake
|
||||||
|
DESTINATION ${CONFIG_FILE_PATH})
|
||||||
|
# 头文件安装命令
|
||||||
|
install(FILES lib/netcdfcpp.h DESTINATION include/netcdfcxx_legacy)
|
||||||
|
install(FILES lib/ncvalues.h DESTINATION include/netcdfcxx_legacy)
|
||||||
|
|
||||||
|
# 以下部分为例子程序的编译
|
||||||
|
# 设置可执行文件的输出地址
|
||||||
|
set(EXECUTABLE_OUTPUT_PATH ${PROJECT_BINARY_DIR}/bin)
|
||||||
|
|
||||||
|
# 例子的编译方法
|
||||||
|
macro(add_sample name)
|
||||||
|
# 添加可执行文件 命令行
|
||||||
|
add_executable(${name} example/${name}.cpp)
|
||||||
|
# 为安装文件添加动态库的搜索地址 在Windows下并没有什么用 直接忽略
|
||||||
|
set_target_properties(${name} PROPERTIES INSTALL_RPATH ${CMAKE_INSTALL_PREFIX}/lib)
|
||||||
|
# 链接动态库
|
||||||
|
target_link_libraries(${name} PUBLIC netcdfcxx_legacy)
|
||||||
|
endmacro()
|
||||||
|
|
||||||
|
add_sample(pres_temp_4D_rd)
|
||||||
|
add_sample(pres_temp_4D_wr)
|
||||||
|
add_sample(sfc_pres_temp_rd)
|
||||||
|
add_sample(sfc_pres_temp_wr)
|
||||||
|
add_sample(simple_xy_rd)
|
||||||
|
add_sample(simple_xy_wr)
|
||||||
126
dep/netcdfcxx_legacy/src/example/pres_temp_4D_rd.cpp
Normal file
@ -0,0 +1,126 @@
|
|||||||
|
/* This is part of the netCDF package.
|
||||||
|
Copyright 2006 University Corporation for Atmospheric Research/Unidata.
|
||||||
|
See COPYRIGHT file for conditions of use.
|
||||||
|
|
||||||
|
This is an example which reads some 4D pressure and temperature
|
||||||
|
values. The data file read by this program is produced by the
|
||||||
|
companion program pres_temp_4D_wr.cpp. It is intended to illustrate
|
||||||
|
the use of the netCDF C++ API.
|
||||||
|
|
||||||
|
This program is part of the netCDF tutorial:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-tutorial
|
||||||
|
|
||||||
|
Full documentation of the netCDF C++ API can be found at:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-cxx
|
||||||
|
|
||||||
|
$Id: pres_temp_4D_rd.cpp,v 1.13 2007/02/14 20:59:21 ed Exp $
|
||||||
|
*/
|
||||||
|
|
||||||
|
#include <iostream>
|
||||||
|
#include "../lib/netcdfcpp.h"
|
||||||
|
|
||||||
|
using namespace std;
|
||||||
|
|
||||||
|
// We are writing 4D data, a 2 x 6 x 12 lvl-lat-lon grid, with 2
|
||||||
|
// timesteps of data.
|
||||||
|
static const int NLVL = 2;
|
||||||
|
static const int NLAT = 6;
|
||||||
|
static const int NLON = 12;
|
||||||
|
static const int NREC = 2;
|
||||||
|
|
||||||
|
// These are used to construct some example data.
|
||||||
|
static const float SAMPLE_PRESSURE = 900.0;
|
||||||
|
static const float SAMPLE_TEMP = 9.0;
|
||||||
|
static const float START_LAT = 25.0;
|
||||||
|
static const float START_LON = -125.0;
|
||||||
|
|
||||||
|
// Return this code to the OS in case of failure.
|
||||||
|
static const int NC_ERR = 2;
|
||||||
|
|
||||||
|
int main()
|
||||||
|
{
|
||||||
|
// These arrays will store the latitude and longitude values.
|
||||||
|
float lats[NLAT], lons[NLON];
|
||||||
|
|
||||||
|
// These arrays will hold the data we will read in. We will only
|
||||||
|
// need enough space to hold one timestep of data; one record.
|
||||||
|
float pres_in[NLVL][NLAT][NLON];
|
||||||
|
float temp_in[NLVL][NLAT][NLON];
|
||||||
|
|
||||||
|
// Change the error behavior of the netCDF C++ API by creating an
|
||||||
|
// NcError object. Until it is destroyed, this NcError object will
|
||||||
|
// ensure that the netCDF C++ API returns error codes on any
|
||||||
|
// failure, prints an error message, and leaves any other error
|
||||||
|
// handling to the calling program. In the case of this example, we
|
||||||
|
// just exit with an NC_ERR error code.
|
||||||
|
NcError err(NcError::verbose_nonfatal);
|
||||||
|
|
||||||
|
// Open the file.
|
||||||
|
NcFile dataFile("pres_temp_4D.nc", NcFile::ReadOnly);
|
||||||
|
|
||||||
|
// Check to see if the file was opened.
|
||||||
|
if(!dataFile.is_valid())
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Get pointers to the latitude and longitude variables.
|
||||||
|
NcVar *latVar, *lonVar;
|
||||||
|
if (!(latVar = dataFile.get_var("latitude")))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(lonVar = dataFile.get_var("longitude")))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Get the lat/lon data from the file.
|
||||||
|
if (!latVar->get(lats, NLAT))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!lonVar->get(lons, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Check the coordinate variable data.
|
||||||
|
for (int lat = 0; lat < NLAT; lat++)
|
||||||
|
if (lats[lat] != START_LAT + 5. * lat)
|
||||||
|
return NC_ERR;
|
||||||
|
for (int lon = 0; lon < NLON; lon++)
|
||||||
|
if (lons[lon] != START_LON + 5. * lon)
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Get pointers to the pressure and temperature variables.
|
||||||
|
NcVar *presVar, *tempVar;
|
||||||
|
if (!(presVar = dataFile.get_var("pressure")))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(tempVar = dataFile.get_var("temperature")))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Read the data. Since we know the contents of the file we know
|
||||||
|
// that the data arrays in this program are the correct size to
|
||||||
|
// hold one timestep.
|
||||||
|
for (int rec = 0; rec < NREC; rec++)
|
||||||
|
{
|
||||||
|
// Read the data one record at a time.
|
||||||
|
if (!presVar->set_cur(rec, 0, 0, 0))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!tempVar->set_cur(rec, 0, 0, 0))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Get 1 record of NLVL by NLAT by NLON values for each variable.
|
||||||
|
if (!presVar->get(&pres_in[0][0][0], 1, NLVL, NLAT, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!tempVar->get(&temp_in[0][0][0], 1, NLVL, NLAT, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Check the data.
|
||||||
|
int i = 0;
|
||||||
|
for (int lvl = 0; lvl < NLVL; lvl++)
|
||||||
|
for (int lat = 0; lat < NLAT; lat++)
|
||||||
|
for (int lon = 0; lon < NLON; lon++)
|
||||||
|
if (pres_in[lvl][lat][lon] != SAMPLE_PRESSURE + i ||
|
||||||
|
temp_in[lvl][lat][lon] != SAMPLE_TEMP + i++)
|
||||||
|
return NC_ERR;
|
||||||
|
} // next record
|
||||||
|
|
||||||
|
// The file is automatically closed by the destructor. This frees
|
||||||
|
// up any internal netCDF resources associated with the file, and
|
||||||
|
// flushes any buffers.
|
||||||
|
|
||||||
|
cout << "*** SUCCESS reading example file pres_temp_4D.nc!" << endl;
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
149
dep/netcdfcxx_legacy/src/example/pres_temp_4D_wr.cpp
Normal file
@ -0,0 +1,149 @@
|
|||||||
|
/* This is part of the netCDF package.
|
||||||
|
Copyright 2006 University Corporation for Atmospheric Research/Unidata.
|
||||||
|
See COPYRIGHT file for conditions of use.
|
||||||
|
|
||||||
|
This is an example program which writes some 4D pressure and
|
||||||
|
temperatures. This example demonstrates the netCDF C++ API.
|
||||||
|
|
||||||
|
This is part of the netCDF tutorial:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-tutorial
|
||||||
|
|
||||||
|
Full documentation of the netCDF C++ API can be found at:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-cxx
|
||||||
|
|
||||||
|
$Id: pres_temp_4D_wr.cpp,v 1.11 2007/01/19 12:52:13 ed Exp $
|
||||||
|
*/
|
||||||
|
|
||||||
|
#include <iostream>
|
||||||
|
#include "../lib/netcdfcpp.h"
|
||||||
|
|
||||||
|
using namespace std;
|
||||||
|
|
||||||
|
// We are writing 4D data, a 2 x 6 x 12 lvl-lat-lon grid, with 2
|
||||||
|
// timesteps of data.
|
||||||
|
static const int NLVL = 2;
|
||||||
|
static const int NLAT = 6;
|
||||||
|
static const int NLON = 12;
|
||||||
|
static const int NREC = 2;
|
||||||
|
|
||||||
|
// These are used to construct some example data.
|
||||||
|
static const float SAMPLE_PRESSURE = 900.0;
|
||||||
|
static const float SAMPLE_TEMP = 9.0;
|
||||||
|
static const float START_LAT = 25.0;
|
||||||
|
static const float START_LON = -125.0;
|
||||||
|
|
||||||
|
// Return this code to the OS in case of failure.
|
||||||
|
static const int NC_ERR = 2;
|
||||||
|
|
||||||
|
int main()
|
||||||
|
{
|
||||||
|
// These arrays will store the latitude and longitude values.
|
||||||
|
float lats[NLAT],lons[NLON];
|
||||||
|
|
||||||
|
// These arrays will hold the data we will write out. We will
|
||||||
|
// only need enough space to hold one timestep of data; one record.
|
||||||
|
float pres_out[NLVL][NLAT][NLON];
|
||||||
|
float temp_out[NLVL][NLAT][NLON];
|
||||||
|
|
||||||
|
int i = 0;
|
||||||
|
|
||||||
|
// Create some pretend data. If this wasn't an example program, we
|
||||||
|
// would have some real data to write for example, model output.
|
||||||
|
for (int lat = 0; lat < NLAT; lat++)
|
||||||
|
lats[lat] = START_LAT + 5. * lat;
|
||||||
|
for (int lon = 0; lon < NLON; lon++)
|
||||||
|
lons[lon] = START_LON + 5. * lon;
|
||||||
|
|
||||||
|
for (int lvl = 0; lvl < NLVL; lvl++)
|
||||||
|
for (int lat = 0; lat < NLAT; lat++)
|
||||||
|
for (int lon = 0; lon < NLON; lon++)
|
||||||
|
{
|
||||||
|
pres_out[lvl][lat][lon] = SAMPLE_PRESSURE + i;
|
||||||
|
temp_out[lvl][lat][lon] = SAMPLE_TEMP + i++;
|
||||||
|
}
|
||||||
|
|
||||||
|
// Change the error behavior of the netCDF C++ API by creating an
|
||||||
|
// NcError object. Until it is destroyed, this NcError object will
|
||||||
|
// ensure that the netCDF C++ API returns error codes on any
|
||||||
|
// failure, prints an error message, and leaves any other error
|
||||||
|
// handling to the calling program. In the case of this example, we
|
||||||
|
// just exit with an NC_ERR error code.
|
||||||
|
NcError err(NcError::verbose_nonfatal);
|
||||||
|
|
||||||
|
// Create the file.
|
||||||
|
NcFile dataFile("pres_temp_4D.nc", NcFile::Replace);
|
||||||
|
|
||||||
|
// Check to see if the file was created.
|
||||||
|
if(!dataFile.is_valid())
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define the dimensions. NetCDF will hand back an ncDim object for
|
||||||
|
// each.
|
||||||
|
NcDim *lvlDim, *latDim, *lonDim, *recDim;
|
||||||
|
if (!(lvlDim = dataFile.add_dim("level", NLVL)))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(latDim = dataFile.add_dim("latitude", NLAT)))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(lonDim = dataFile.add_dim("longitude", NLON)))
|
||||||
|
return NC_ERR;
|
||||||
|
// Add an unlimited dimension...
|
||||||
|
if (!(recDim = dataFile.add_dim("time", NREC)))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define the coordinate variables.
|
||||||
|
NcVar *latVar, *lonVar;
|
||||||
|
if (!(latVar = dataFile.add_var("latitude", ncFloat, latDim)))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(lonVar = dataFile.add_var("longitude", ncFloat, lonDim)))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define units attributes for coordinate vars. This attaches a
|
||||||
|
// text attribute to each of the coordinate variables, containing
|
||||||
|
// the units.
|
||||||
|
if (!latVar->add_att("units", "degrees_north"))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!lonVar->add_att("units", "degrees_east"))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define the netCDF variables for the pressure and temperature
|
||||||
|
// data.
|
||||||
|
NcVar *presVar, *tempVar;
|
||||||
|
if (!(presVar = dataFile.add_var("pressure", ncFloat, recDim,
|
||||||
|
lvlDim, latDim, lonDim)))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(tempVar = dataFile.add_var("temperature", ncFloat, recDim,
|
||||||
|
lvlDim, latDim, lonDim)))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define units attributes for data variables.
|
||||||
|
if (!presVar->add_att("units", "hPa"))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!tempVar->add_att("units", "celsius"))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Write the coordinate variable data to the file.
|
||||||
|
if (!latVar->put(lats, NLAT))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!lonVar->put(lons, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Write the pretend data. This will write our surface pressure and
|
||||||
|
// surface temperature data. The arrays only hold one timestep
|
||||||
|
// worth of data. We will just rewrite the same data for each
|
||||||
|
// timestep. In a real application, the data would change between
|
||||||
|
// timesteps.
|
||||||
|
for (int rec = 0; rec < NREC; rec++)
|
||||||
|
{
|
||||||
|
if (!presVar->put_rec(&pres_out[0][0][0], rec))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!tempVar->put_rec(&temp_out[0][0][0], rec))
|
||||||
|
return NC_ERR;
|
||||||
|
}
|
||||||
|
|
||||||
|
// The file is automatically closed by the destructor. This frees
|
||||||
|
// up any internal netCDF resources associated with the file, and
|
||||||
|
// flushes any buffers.
|
||||||
|
|
||||||
|
cout << "*** SUCCESS writing example file pres_temp_4D.nc!" << endl;
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
161
dep/netcdfcxx_legacy/src/example/sfc_pres_temp_rd.cpp
Normal file
@ -0,0 +1,161 @@
|
|||||||
|
/* This is part of the netCDF package.
|
||||||
|
Copyright 2006 University Corporation for Atmospheric Research/Unidata.
|
||||||
|
See COPYRIGHT file for conditions of use.
|
||||||
|
|
||||||
|
This is an example which reads some surface pressure and
|
||||||
|
temperatures. The data file read by this program is produced
|
||||||
|
companion program sfc_pres_temp_wr.cxx. It is intended to
|
||||||
|
illustrate the use of the netCDF C++ API.
|
||||||
|
|
||||||
|
This program is part of the netCDF tutorial:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-tutorial
|
||||||
|
|
||||||
|
Full documentation of the netCDF C++ API can be found at:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-cxx
|
||||||
|
|
||||||
|
$Id: sfc_pres_temp_rd.cpp,v 1.17 2008/05/16 13:42:28 ed Exp $
|
||||||
|
*/
|
||||||
|
|
||||||
|
#include <iostream>
|
||||||
|
#include <cstring>
|
||||||
|
#include "../lib/netcdfcpp.h"
|
||||||
|
|
||||||
|
using namespace std;
|
||||||
|
|
||||||
|
// We are reading 2D data, a 6 x 12 lat-lon grid.
|
||||||
|
static const int NLAT = 6;
|
||||||
|
static const int NLON = 12;
|
||||||
|
|
||||||
|
// These are used to calculate the values we expect to find.
|
||||||
|
static const float SAMPLE_PRESSURE = 900;
|
||||||
|
static const float SAMPLE_TEMP = 9.0;
|
||||||
|
static const float START_LAT = 25.0;
|
||||||
|
static const float START_LON = -125.0;
|
||||||
|
|
||||||
|
// Return this code to the OS in case of failure.
|
||||||
|
static const int NC_ERR = 2;
|
||||||
|
|
||||||
|
int main(void)
|
||||||
|
{
|
||||||
|
// These will hold our pressure and temperature data.
|
||||||
|
float presIn[NLAT][NLON];
|
||||||
|
float tempIn[NLAT][NLON];
|
||||||
|
|
||||||
|
// These will hold our latitudes and longitudes.
|
||||||
|
float latsIn[NLAT];
|
||||||
|
float lonsIn[NLON];
|
||||||
|
|
||||||
|
// Change the error behavior of the netCDF C++ API by creating an
|
||||||
|
// NcError object. Until it is destroyed, this NcError object will
|
||||||
|
// ensure that the netCDF C++ API silently returns error codes on
|
||||||
|
// any failure, and leaves any other error handling to the calling
|
||||||
|
// program. In the case of this example, we just exit with an
|
||||||
|
// NC_ERR error code.
|
||||||
|
NcError err(NcError::silent_nonfatal);
|
||||||
|
|
||||||
|
// Open the file and check to make sure it's valid.
|
||||||
|
NcFile dataFile("sfc_pres_temp.nc", NcFile::ReadOnly);
|
||||||
|
if(!dataFile.is_valid())
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// There are a number of inquiry functions in netCDF which can be
|
||||||
|
// used to learn about an unknown netCDF file. In this case we know
|
||||||
|
// that there are 2 netCDF dimensions, 4 netCDF variables, no
|
||||||
|
// global attributes, and no unlimited dimension.
|
||||||
|
if (dataFile.num_dims() != 2 || dataFile.num_vars() != 4 ||
|
||||||
|
dataFile.num_atts() != 0 || dataFile.rec_dim() != 0)
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// We get back a pointer to each NcVar we request. Get the
|
||||||
|
// latitude and longitude coordinate variables.
|
||||||
|
NcVar *latVar, *lonVar;
|
||||||
|
if (!(latVar = dataFile.get_var("latitude")))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(lonVar = dataFile.get_var("longitude")))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Read the latitude and longitude coordinate variables into arrays
|
||||||
|
// latsIn and lonsIn.
|
||||||
|
if (!latVar->get(latsIn, NLAT))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!lonVar->get(lonsIn, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Check the coordinate variable data.
|
||||||
|
for(int lat = 0; lat < NLAT; lat++)
|
||||||
|
if (latsIn[lat] != START_LAT + 5. * lat)
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Check longitude values.
|
||||||
|
for (int lon = 0; lon < NLON; lon++)
|
||||||
|
if (lonsIn[lon] != START_LON + 5. * lon)
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// We get back a pointer to each NcVar we request.
|
||||||
|
NcVar *presVar, *tempVar;
|
||||||
|
if (!(presVar = dataFile.get_var("pressure")))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(tempVar = dataFile.get_var("temperature")))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Read the data. Since we know the contents of the file we know
|
||||||
|
// that the data arrays in this program are the correct size to
|
||||||
|
// hold all the data.
|
||||||
|
if (!presVar->get(&presIn[0][0], NLAT, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!tempVar->get(&tempIn[0][0], NLAT, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Check the data.
|
||||||
|
for (int lat = 0; lat < NLAT; lat++)
|
||||||
|
for (int lon = 0; lon < NLON; lon++)
|
||||||
|
if (presIn[lat][lon] != SAMPLE_PRESSURE + (lon * NLAT + lat)
|
||||||
|
|| tempIn[lat][lon] != SAMPLE_TEMP + .25 * (lon * NLAT + lat))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Each of the netCDF variables has a "units" attribute. Let's read
|
||||||
|
// them and check them.
|
||||||
|
NcAtt *att;
|
||||||
|
char *units;
|
||||||
|
|
||||||
|
if (!(att = latVar->get_att("units")))
|
||||||
|
return NC_ERR;
|
||||||
|
units = att->as_string(0);
|
||||||
|
if (strncmp(units, "degrees_north", strlen("degrees_north")))
|
||||||
|
return NC_ERR;
|
||||||
|
// Attributes and attribute values should be deleted by the caller
|
||||||
|
// when no longer needed, to prevent memory leaks.
|
||||||
|
delete units;
|
||||||
|
delete att;
|
||||||
|
|
||||||
|
if (!(att = lonVar->get_att("units")))
|
||||||
|
return NC_ERR;
|
||||||
|
units = att->as_string(0);
|
||||||
|
if (strncmp(units, "degrees_east", strlen("degrees_east")))
|
||||||
|
return NC_ERR;
|
||||||
|
delete units;
|
||||||
|
delete att;
|
||||||
|
|
||||||
|
if (!(att = presVar->get_att("units")))
|
||||||
|
return NC_ERR;
|
||||||
|
units = att->as_string(0);
|
||||||
|
if (strncmp(units, "hPa", strlen("hPa")))
|
||||||
|
return NC_ERR;
|
||||||
|
delete units;
|
||||||
|
delete att;
|
||||||
|
|
||||||
|
if (!(att = tempVar->get_att("units")))
|
||||||
|
return NC_ERR;
|
||||||
|
units = att->as_string(0);
|
||||||
|
if (strncmp(units, "celsius", strlen("celsius")))
|
||||||
|
return NC_ERR;
|
||||||
|
delete units;
|
||||||
|
delete att;
|
||||||
|
|
||||||
|
// The file will be automatically closed by the destructor. This
|
||||||
|
// frees up any internal netCDF resources associated with the file,
|
||||||
|
// and flushes any buffers.
|
||||||
|
cout << "*** SUCCESS reading example file sfc_pres_temp.nc!" << endl;
|
||||||
|
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
142
dep/netcdfcxx_legacy/src/example/sfc_pres_temp_wr.cpp
Normal file
@ -0,0 +1,142 @@
|
|||||||
|
/* This is part of the netCDF package.
|
||||||
|
Copyright 2006 University Corporation for Atmospheric Research/Unidata.
|
||||||
|
See COPYRIGHT file for conditions of use.
|
||||||
|
|
||||||
|
This example writes some surface pressure and temperatures. It is
|
||||||
|
intended to illustrate the use of the netCDF C++ API. The companion
|
||||||
|
program sfc_pres_temp_rd.cpp shows how to read the netCDF data file
|
||||||
|
created by this program.
|
||||||
|
|
||||||
|
This program is part of the netCDF tutorial:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-tutorial
|
||||||
|
|
||||||
|
Full documentation of the netCDF C++ API can be found at:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-cxx
|
||||||
|
|
||||||
|
$Id: sfc_pres_temp_wr.cpp,v 1.12 2007/01/19 12:52:13 ed Exp $
|
||||||
|
*/
|
||||||
|
|
||||||
|
#include <iostream>
|
||||||
|
#include "../lib/netcdfcpp.h"
|
||||||
|
|
||||||
|
using namespace std;
|
||||||
|
|
||||||
|
// We are writing 2D data, a 6 x 12 lat-lon grid. We will need two
|
||||||
|
// netCDF dimensions.
|
||||||
|
static const int NLAT = 6;
|
||||||
|
static const int NLON = 12;
|
||||||
|
|
||||||
|
// These are used to construct some example data.
|
||||||
|
static const float SAMPLE_PRESSURE = 900;
|
||||||
|
static const float SAMPLE_TEMP = 9.0;
|
||||||
|
static const float START_LAT = 25.0;
|
||||||
|
static const float START_LON = -125.0;
|
||||||
|
|
||||||
|
// Return this to OS if there is a failure.
|
||||||
|
static const int NC_ERR = 2;
|
||||||
|
|
||||||
|
int main(void)
|
||||||
|
{
|
||||||
|
// These will hold our pressure and temperature data.
|
||||||
|
float presOut[NLAT][NLON];
|
||||||
|
float tempOut[NLAT][NLON];
|
||||||
|
|
||||||
|
// These will hold our latitudes and longitudes.
|
||||||
|
float lats[NLAT];
|
||||||
|
float lons[NLON];
|
||||||
|
|
||||||
|
// Create some pretend data. If this wasn't an example program, we
|
||||||
|
// would have some real data to write, for example, model
|
||||||
|
// output.
|
||||||
|
for(int lat = 0; lat < NLAT; lat++)
|
||||||
|
lats[lat] = START_LAT + 5. * lat;
|
||||||
|
|
||||||
|
for(int lon = 0; lon < NLON; lon++)
|
||||||
|
lons[lon] = START_LON + 5. * lon;
|
||||||
|
|
||||||
|
for (int lat = 0; lat < NLAT; lat++)
|
||||||
|
for(int lon = 0; lon < NLON; lon++)
|
||||||
|
{
|
||||||
|
presOut[lat][lon] = SAMPLE_PRESSURE + (lon * NLAT + lat);
|
||||||
|
tempOut[lat][lon] = SAMPLE_TEMP + .25 * (lon * NLAT + lat);
|
||||||
|
}
|
||||||
|
|
||||||
|
// Change the error behavior of the netCDF C++ API by creating an
|
||||||
|
// NcError object. Until it is destroyed, this NcError object will
|
||||||
|
// ensure that the netCDF C++ API silently returns error codes
|
||||||
|
// on any failure, and leaves any other error handling to the
|
||||||
|
// calling program. In the case of this example, we just exit with
|
||||||
|
// an NC_ERR error code.
|
||||||
|
NcError err(NcError::silent_nonfatal);
|
||||||
|
|
||||||
|
// Create the file. The Replace parameter tells netCDF to overwrite
|
||||||
|
// this file, if it already exists.
|
||||||
|
NcFile dataFile("sfc_pres_temp.nc", NcFile::Replace);
|
||||||
|
|
||||||
|
// Check to see if the file was created.
|
||||||
|
if(!dataFile.is_valid())
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define the dimensions. NetCDF will hand back an ncDim object for
|
||||||
|
// each.
|
||||||
|
NcDim *latDim, *lonDim;
|
||||||
|
if (!(latDim = dataFile.add_dim("latitude", NLAT)))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(lonDim = dataFile.add_dim("longitude", NLON)))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// In addition to the latitude and longitude dimensions, we will
|
||||||
|
// also create latitude and longitude netCDF variables which will
|
||||||
|
// hold the actual latitudes and longitudes. Since they hold data
|
||||||
|
// about the coordinate system, the netCDF term for these is:
|
||||||
|
// "coordinate variables."
|
||||||
|
NcVar *latVar, *lonVar;
|
||||||
|
if (!(latVar = dataFile.add_var("latitude", ncFloat, latDim)))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(lonVar = dataFile.add_var("longitude", ncFloat, lonDim)))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define units attributes for coordinate vars. This attaches a
|
||||||
|
// text attribute to each of the coordinate variables, containing
|
||||||
|
// the units.
|
||||||
|
if (!lonVar->add_att("units", "degrees_east"))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!latVar->add_att("units", "degrees_north"))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define the netCDF data variables.
|
||||||
|
NcVar *presVar, *tempVar;
|
||||||
|
if (!(presVar = dataFile.add_var("pressure", ncFloat, latDim, lonDim)))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!(tempVar = dataFile.add_var("temperature", ncFloat, latDim, lonDim)))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Define units attributes for variables.
|
||||||
|
if (!presVar->add_att("units", "hPa"))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!tempVar->add_att("units", "celsius"))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Write the coordinate variable data. This will put the latitudes
|
||||||
|
// and longitudes of our data grid into the netCDF file.
|
||||||
|
if (!latVar->put(lats, NLAT))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!lonVar->put(lons, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// Write the pretend data. This will write our surface pressure and
|
||||||
|
// surface temperature data. The arrays of data are the same size
|
||||||
|
// as the netCDF variables we have defined, and below we write them
|
||||||
|
// each in one step.
|
||||||
|
if (!presVar->put(&presOut[0][0], NLAT, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
if (!tempVar->put(&tempOut[0][0], NLAT, NLON))
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// The file is automatically closed by the destructor. This frees
|
||||||
|
// up any internal netCDF resources associated with the file, and
|
||||||
|
// flushes any buffers.
|
||||||
|
cout << "*** SUCCESS writing example file sfc_pres_temp.nc!" << endl;
|
||||||
|
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
67
dep/netcdfcxx_legacy/src/example/simple_xy_rd.cpp
Normal file
@ -0,0 +1,67 @@
|
|||||||
|
/* This is part of the netCDF package.
|
||||||
|
Copyright 2006 University Corporation for Atmospheric Research/Unidata.
|
||||||
|
See COPYRIGHT file for conditions of use.
|
||||||
|
|
||||||
|
This is a very simple example which reads a 2D array of
|
||||||
|
sample data produced by simple_xy_wr.cpp.
|
||||||
|
|
||||||
|
This example is part of the netCDF tutorial:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-tutorial
|
||||||
|
|
||||||
|
Full documentation of the netCDF C++ API can be found at:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-cxx
|
||||||
|
|
||||||
|
$Id: simple_xy_rd.cpp,v 1.13 2007/01/19 12:52:13 ed Exp $
|
||||||
|
*/
|
||||||
|
|
||||||
|
#include <iostream>
|
||||||
|
#include "../lib/netcdfcpp.h"
|
||||||
|
|
||||||
|
using namespace std;
|
||||||
|
|
||||||
|
// We are reading 2D data, a 6 x 12 grid.
|
||||||
|
static const int NX = 6;
|
||||||
|
static const int NY = 12;
|
||||||
|
|
||||||
|
// Return this in event of a problem.
|
||||||
|
static const int NC_ERR = 2;
|
||||||
|
|
||||||
|
int main(void)
|
||||||
|
{
|
||||||
|
// This is the array we will read.
|
||||||
|
int dataIn[NX][NY];
|
||||||
|
|
||||||
|
// Open the file. The ReadOnly parameter tells netCDF we want
|
||||||
|
// read-only access to the file.
|
||||||
|
NcFile dataFile("simple_xy.nc", NcFile::ReadOnly);
|
||||||
|
|
||||||
|
// You should always check whether a netCDF file open or creation
|
||||||
|
// constructor succeeded.
|
||||||
|
if (!dataFile.is_valid())
|
||||||
|
{
|
||||||
|
cout << "Couldn't open file!\n";
|
||||||
|
return NC_ERR;
|
||||||
|
}
|
||||||
|
|
||||||
|
// For other method calls, the default behavior of the C++ API is
|
||||||
|
// to exit with a message if there is an error. If that behavior
|
||||||
|
// is OK, there is no need to check return values in simple cases
|
||||||
|
// like the following.
|
||||||
|
|
||||||
|
// Retrieve the variable named "data"
|
||||||
|
NcVar *data = dataFile.get_var("data");
|
||||||
|
|
||||||
|
// Read all the values from the "data" variable into memory.
|
||||||
|
data->get(&dataIn[0][0], NX, NY);
|
||||||
|
|
||||||
|
// Check the values.
|
||||||
|
for (int i = 0; i < NX; i++)
|
||||||
|
for (int j = 0; j < NY; j++)
|
||||||
|
if (dataIn[i][j] != i * NY + j)
|
||||||
|
return NC_ERR;
|
||||||
|
|
||||||
|
// The netCDF file is automatically closed by the NcFile destructor
|
||||||
|
cout << "*** SUCCESS reading example file simple_xy.nc!" << endl;
|
||||||
|
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
80
dep/netcdfcxx_legacy/src/example/simple_xy_wr.cpp
Normal file
@ -0,0 +1,80 @@
|
|||||||
|
/* This is part of the netCDF package.
|
||||||
|
Copyright 2006 University Corporation for Atmospheric Research/Unidata.
|
||||||
|
See COPYRIGHT file for conditions of use.
|
||||||
|
|
||||||
|
This is a very simple example which writes a 2D array of
|
||||||
|
sample data. To handle this in netCDF we create two shared
|
||||||
|
dimensions, "x" and "y", and a netCDF variable, called "data".
|
||||||
|
|
||||||
|
This example is part of the netCDF tutorial:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-tutorial
|
||||||
|
|
||||||
|
Full documentation of the netCDF C++ API can be found at:
|
||||||
|
http://www.unidata.ucar.edu/software/netcdf/docs/netcdf-cxx
|
||||||
|
|
||||||
|
$Id: simple_xy_wr.cpp,v 1.15 2007/01/19 12:52:13 ed Exp $
|
||||||
|
*/
|
||||||
|
|
||||||
|
#include <iostream>
|
||||||
|
#include "../lib/netcdfcpp.h"
|
||||||
|
|
||||||
|
using namespace std;
|
||||||
|
|
||||||
|
// We are writing 2D data, a 6 x 12 grid.
|
||||||
|
static const int NX = 6;
|
||||||
|
static const int NY = 12;
|
||||||
|
|
||||||
|
// Return this in event of a problem.
|
||||||
|
static const int NC_ERR = 2;
|
||||||
|
|
||||||
|
int
|
||||||
|
main(void)
|
||||||
|
{
|
||||||
|
// This is the data array we will write. It will just be filled
|
||||||
|
// with a progression of numbers for this example.
|
||||||
|
int dataOut[NX][NY];
|
||||||
|
|
||||||
|
// Create some pretend data. If this wasn't an example program, we
|
||||||
|
// would have some real data to write, for example, model output.
|
||||||
|
for(int i = 0; i < NX; i++)
|
||||||
|
for(int j = 0; j < NY; j++)
|
||||||
|
dataOut[i][j] = i * NY + j;
|
||||||
|
|
||||||
|
// Create the file. The Replace parameter tells netCDF to overwrite
|
||||||
|
// this file, if it already exists.
|
||||||
|
NcFile dataFile("simple_xy.nc", NcFile::Replace);
|
||||||
|
|
||||||
|
// You should always check whether a netCDF file creation or open
|
||||||
|
// constructor succeeded.
|
||||||
|
if (!dataFile.is_valid())
|
||||||
|
{
|
||||||
|
cout << "Couldn't open file!\n";
|
||||||
|
return NC_ERR;
|
||||||
|
}
|
||||||
|
|
||||||
|
// For other method calls, the default behavior of the C++ API is
|
||||||
|
// to exit with a message if there is an error. If that behavior
|
||||||
|
// is OK, there is no need to check return values in simple cases
|
||||||
|
// like the following.
|
||||||
|
|
||||||
|
// When we create netCDF dimensions, we get back a pointer to an
|
||||||
|
// NcDim for each one.
|
||||||
|
NcDim* xDim = dataFile.add_dim("x", NX);
|
||||||
|
NcDim* yDim = dataFile.add_dim("y", NY);
|
||||||
|
|
||||||
|
// Define a netCDF variable. The type of the variable in this case
|
||||||
|
// is ncInt (32-bit integer).
|
||||||
|
NcVar *data = dataFile.add_var("data", ncInt, xDim, yDim);
|
||||||
|
|
||||||
|
// Write the pretend data to the file. Although netCDF supports
|
||||||
|
// reading and writing subsets of data, in this case we write all
|
||||||
|
// the data in one operation.
|
||||||
|
data->put(&dataOut[0][0], NX, NY);
|
||||||
|
|
||||||
|
// The file will be automatically close when the NcFile object goes
|
||||||
|
// out of scope. This frees up any internal netCDF resources
|
||||||
|
// associated with the file, and flushes any buffers.
|
||||||
|
cout << "*** SUCCESS writing example file simple_xy.nc!" << endl;
|
||||||
|
|
||||||
|
return 0;
|
||||||
|
}
|
||||||
332
dep/netcdfcxx_legacy/src/lib/ncvalues.cpp
Normal file
@ -0,0 +1,332 @@
|
|||||||
|
/*********************************************************************
|
||||||
|
* Copyright 1992, University Corporation for Atmospheric Research
|
||||||
|
* See netcdf/README file for copying and redistribution conditions.
|
||||||
|
*
|
||||||
|
* Purpose: implementation of classes of typed arrays for netCDF
|
||||||
|
*
|
||||||
|
* $Header: /upc/share/CVS/netcdf-3/cxx/ncvalues.cpp,v 1.12 2008/03/05 16:45:32 russ Exp $
|
||||||
|
*********************************************************************/
|
||||||
|
|
||||||
|
#include <iostream>
|
||||||
|
#include <string>
|
||||||
|
#include <cstring>
|
||||||
|
|
||||||
|
#include "netcdf_config.h"
|
||||||
|
#include "ncvalues.h"
|
||||||
|
#pragma warning(disable : 4996)
|
||||||
|
|
||||||
|
NcValues::NcValues( void ) : the_type(ncNoType), the_number(0)
|
||||||
|
{}
|
||||||
|
|
||||||
|
NcValues::NcValues(NcType type, long num)
|
||||||
|
: the_type(type), the_number(num)
|
||||||
|
{}
|
||||||
|
|
||||||
|
NcValues::~NcValues( void )
|
||||||
|
{}
|
||||||
|
|
||||||
|
long NcValues::num( void )
|
||||||
|
{
|
||||||
|
return the_number;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& operator<< (std::ostream& os, const NcValues& vals)
|
||||||
|
{
|
||||||
|
return vals.print(os);
|
||||||
|
}
|
||||||
|
|
||||||
|
implement(NcValues,ncbyte)
|
||||||
|
implement(NcValues,char)
|
||||||
|
implement(NcValues,short)
|
||||||
|
implement(NcValues,int)
|
||||||
|
implement(NcValues,nclong)
|
||||||
|
implement(NcValues,long)
|
||||||
|
implement(NcValues,float)
|
||||||
|
implement(NcValues,double)
|
||||||
|
|
||||||
|
Ncbytes_for_one_implement(ncbyte)
|
||||||
|
Ncbytes_for_one_implement(char)
|
||||||
|
Ncbytes_for_one_implement(short)
|
||||||
|
Ncbytes_for_one_implement(int)
|
||||||
|
Ncbytes_for_one_implement(nclong)
|
||||||
|
Ncbytes_for_one_implement(long)
|
||||||
|
Ncbytes_for_one_implement(float)
|
||||||
|
Ncbytes_for_one_implement(double)
|
||||||
|
|
||||||
|
as_ncbyte_implement(short)
|
||||||
|
as_ncbyte_implement(int)
|
||||||
|
as_ncbyte_implement(nclong)
|
||||||
|
as_ncbyte_implement(long)
|
||||||
|
as_ncbyte_implement(float)
|
||||||
|
as_ncbyte_implement(double)
|
||||||
|
|
||||||
|
inline ncbyte NcValues_char::as_ncbyte( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline ncbyte NcValues_ncbyte::as_ncbyte( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
as_char_implement(short)
|
||||||
|
as_char_implement(int)
|
||||||
|
as_char_implement(nclong)
|
||||||
|
as_char_implement(long)
|
||||||
|
as_char_implement(float)
|
||||||
|
as_char_implement(double)
|
||||||
|
|
||||||
|
inline char NcValues_ncbyte::as_char( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n] > CHAR_MAX ? ncBad_char : (char) the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline char NcValues_char::as_char( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
as_short_implement(int)
|
||||||
|
as_short_implement(nclong)
|
||||||
|
as_short_implement(long)
|
||||||
|
as_short_implement(float)
|
||||||
|
as_short_implement(double)
|
||||||
|
|
||||||
|
inline short NcValues_ncbyte::as_short( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline short NcValues_char::as_short( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline short NcValues_short::as_short( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
as_int_implement(float)
|
||||||
|
as_int_implement(double)
|
||||||
|
|
||||||
|
inline int NcValues_ncbyte::as_int( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline int NcValues_char::as_int( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline int NcValues_short::as_int( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline int NcValues_int::as_int( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline int NcValues_nclong::as_int( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline int NcValues_long::as_int( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
as_nclong_implement(float)
|
||||||
|
as_nclong_implement(double)
|
||||||
|
|
||||||
|
inline nclong NcValues_ncbyte::as_nclong( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline nclong NcValues_char::as_nclong( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline nclong NcValues_short::as_nclong( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline nclong NcValues_int::as_nclong( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline nclong NcValues_nclong::as_nclong( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline nclong NcValues_long::as_nclong( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
as_long_implement(float)
|
||||||
|
as_long_implement(double)
|
||||||
|
|
||||||
|
inline long NcValues_ncbyte::as_long( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline long NcValues_char::as_long( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline long NcValues_short::as_long( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline long NcValues_int::as_long( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline long NcValues_nclong::as_long( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
inline long NcValues_long::as_long( long n ) const
|
||||||
|
{
|
||||||
|
return the_values[n];
|
||||||
|
}
|
||||||
|
|
||||||
|
as_float_implement(ncbyte)
|
||||||
|
as_float_implement(char)
|
||||||
|
as_float_implement(short)
|
||||||
|
as_float_implement(int)
|
||||||
|
as_float_implement(nclong)
|
||||||
|
as_float_implement(long)
|
||||||
|
as_float_implement(float)
|
||||||
|
as_float_implement(double)
|
||||||
|
|
||||||
|
as_double_implement(ncbyte)
|
||||||
|
as_double_implement(char)
|
||||||
|
as_double_implement(short)
|
||||||
|
as_double_implement(int)
|
||||||
|
as_double_implement(nclong)
|
||||||
|
as_double_implement(long)
|
||||||
|
as_double_implement(float)
|
||||||
|
as_double_implement(double)
|
||||||
|
|
||||||
|
as_string_implement(short)
|
||||||
|
as_string_implement(int)
|
||||||
|
as_string_implement(nclong)
|
||||||
|
as_string_implement(long)
|
||||||
|
as_string_implement(float)
|
||||||
|
as_string_implement(double)
|
||||||
|
|
||||||
|
inline char* NcValues_ncbyte::as_string( long n ) const
|
||||||
|
{
|
||||||
|
char* s = new char[the_number + 1];
|
||||||
|
s[the_number] = '\0';
|
||||||
|
strncpy(s, (const char*)the_values + n, (int)the_number);
|
||||||
|
return s;
|
||||||
|
}
|
||||||
|
|
||||||
|
inline char* NcValues_char::as_string( long n ) const
|
||||||
|
{
|
||||||
|
char* s = new char[the_number + 1];
|
||||||
|
s[the_number] = '\0';
|
||||||
|
strncpy(s, (const char*)the_values + n, (int)the_number);
|
||||||
|
return s;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& NcValues_short::print(std::ostream& os) const
|
||||||
|
{
|
||||||
|
for(int i = 0; i < the_number - 1; i++)
|
||||||
|
os << the_values[i] << ", ";
|
||||||
|
if (the_number > 0)
|
||||||
|
os << the_values[the_number-1] ;
|
||||||
|
return os;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& NcValues_int::print(std::ostream& os) const
|
||||||
|
{
|
||||||
|
for(int i = 0; i < the_number - 1; i++)
|
||||||
|
os << the_values[i] << ", ";
|
||||||
|
if (the_number > 0)
|
||||||
|
os << the_values[the_number-1] ;
|
||||||
|
return os;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& NcValues_nclong::print(std::ostream& os) const
|
||||||
|
{
|
||||||
|
for(int i = 0; i < the_number - 1; i++)
|
||||||
|
os << the_values[i] << ", ";
|
||||||
|
if (the_number > 0)
|
||||||
|
os << the_values[the_number-1] ;
|
||||||
|
return os;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& NcValues_long::print(std::ostream& os) const
|
||||||
|
{
|
||||||
|
for(int i = 0; i < the_number - 1; i++)
|
||||||
|
os << the_values[i] << ", ";
|
||||||
|
if (the_number > 0)
|
||||||
|
os << the_values[the_number-1] ;
|
||||||
|
return os;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& NcValues_ncbyte::print(std::ostream& os) const
|
||||||
|
{
|
||||||
|
for(int i = 0; i < the_number - 1; i++)
|
||||||
|
os << the_values[i] << ", ";
|
||||||
|
if (the_number > 0)
|
||||||
|
os << the_values[the_number-1] ;
|
||||||
|
return os;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& NcValues_char::print(std::ostream& os) const
|
||||||
|
{
|
||||||
|
os << '"';
|
||||||
|
long len = the_number;
|
||||||
|
while (the_values[--len] == '\0') // don't output trailing null bytes
|
||||||
|
;
|
||||||
|
for(int i = 0; i <= len; i++)
|
||||||
|
os << the_values[i] ;
|
||||||
|
os << '"';
|
||||||
|
|
||||||
|
return os;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& NcValues_float::print(std::ostream& os) const
|
||||||
|
{
|
||||||
|
std::streamsize save=os.precision();
|
||||||
|
os.precision(7);
|
||||||
|
for(int i = 0; i < the_number - 1; i++)
|
||||||
|
os << the_values[i] << ", ";
|
||||||
|
if (the_number > 0)
|
||||||
|
os << the_values[the_number-1] ;
|
||||||
|
os.precision(save);
|
||||||
|
return os;
|
||||||
|
}
|
||||||
|
|
||||||
|
std::ostream& NcValues_double::print(std::ostream& os) const
|
||||||
|
{
|
||||||
|
std::streamsize save=os.precision();
|
||||||
|
os.precision(15);
|
||||||
|
for(int i = 0; i < the_number - 1; i++)
|
||||||
|
os << the_values[i] << ", ";
|
||||||
|
if (the_number > 0)
|
||||||
|
os << the_values[the_number-1];
|
||||||
|
os.precision(save);
|
||||||
|
return os;
|
||||||
|
}
|
||||||
279
dep/netcdfcxx_legacy/src/lib/ncvalues.h
Normal file
@ -0,0 +1,279 @@
|
|||||||
|
/*********************************************************************
|
||||||
|
* Copyright 1992, University Corporation for Atmospheric Research
|
||||||
|
* See netcdf/README file for copying and redistribution conditions.
|
||||||
|
*
|
||||||
|
* Purpose: interface for classes of typed arrays for netCDF
|
||||||
|
*
|
||||||
|
* $Header: /upc/share/CVS/netcdf-3/cxx/ncvalues.h,v 1.7 2006/07/26 21:12:06 russ Exp $
|
||||||
|
*********************************************************************/
|
||||||
|
|
||||||
|
#ifndef Ncvalues_def
|
||||||
|
#define Ncvalues_def
|
||||||
|
|
||||||
|
#include <iostream>
|
||||||
|
#include <sstream>
|
||||||
|
#include <limits.h>
|
||||||
|
#include "netcdf.h"
|
||||||
|
|
||||||
|
// Documentation warned this might change and now it has, for
|
||||||
|
// consistency with C interface
|
||||||
|
typedef signed char ncbyte;
|
||||||
|
|
||||||
|
#define NC_UNSPECIFIED ((nc_type)0)
|
||||||
|
|
||||||
|
// C++ interface dates from before netcdf-3, still uses some netcdf-2 names
|
||||||
|
#ifdef NO_NETCDF_2
|
||||||
|
#define NC_LONG NC_INT
|
||||||
|
#define FILL_LONG NC_FILL_INT
|
||||||
|
typedef int nclong;
|
||||||
|
#define NC_FATAL 1
|
||||||
|
#define NC_VERBOSE 2
|
||||||
|
#endif
|
||||||
|
|
||||||
|
enum NcType
|
||||||
|
{
|
||||||
|
ncNoType = NC_UNSPECIFIED,
|
||||||
|
ncByte = NC_BYTE,
|
||||||
|
ncChar = NC_CHAR,
|
||||||
|
ncShort = NC_SHORT,
|
||||||
|
ncInt = NC_INT,
|
||||||
|
ncLong = NC_LONG, // deprecated, someday want to use for 64-bit ints
|
||||||
|
ncFloat = NC_FLOAT,
|
||||||
|
ncDouble = NC_DOUBLE
|
||||||
|
};
|
||||||
|
|
||||||
|
#define ncBad_ncbyte ncBad_byte
|
||||||
|
static const ncbyte ncBad_byte = NC_FILL_BYTE;
|
||||||
|
static const char ncBad_char = NC_FILL_CHAR;
|
||||||
|
static const short ncBad_short = NC_FILL_SHORT;
|
||||||
|
static const nclong ncBad_nclong = FILL_LONG; // deprecated
|
||||||
|
static const int ncBad_int = NC_FILL_INT;
|
||||||
|
static const long ncBad_long = FILL_LONG; // deprecated
|
||||||
|
static const float ncBad_float = NC_FILL_FLOAT;
|
||||||
|
static const double ncBad_double = NC_FILL_DOUBLE;
|
||||||
|
|
||||||
|
// macros to glue tokens together to form new names (used to be in generic.h)
|
||||||
|
#define name2(a,b) a ## b
|
||||||
|
#define declare(clas,t) name2(clas,declare)(t)
|
||||||
|
#define implement(clas,t) name2(clas,implement)(t)
|
||||||
|
// This is the same as the name2 macro, but we need to define our own
|
||||||
|
// version since rescanning something generated with the name2 macro
|
||||||
|
// won't necessarily cause name2 to be expanded again.
|
||||||
|
#define makename2(z, y) makename2_x(z, y)
|
||||||
|
#define makename2_x(z, y) z##y
|
||||||
|
|
||||||
|
#define NcVal(TYPE) makename2(NcValues_,TYPE)
|
||||||
|
|
||||||
|
#define NcValuesdeclare(TYPE) \
|
||||||
|
class NcVal(TYPE) : public NcValues \
|
||||||
|
{ \
|
||||||
|
public: \
|
||||||
|
NcVal(TYPE)( void ); \
|
||||||
|
NcVal(TYPE)(long num); \
|
||||||
|
NcVal(TYPE)(long num, const TYPE* vals); \
|
||||||
|
NcVal(TYPE)(const NcVal(TYPE)&); \
|
||||||
|
virtual NcVal(TYPE)& operator=(const NcVal(TYPE)&); \
|
||||||
|
virtual ~NcVal(TYPE)( void ); \
|
||||||
|
virtual void* base( void ) const; \
|
||||||
|
virtual int bytes_for_one( void ) const; \
|
||||||
|
virtual ncbyte as_ncbyte( long n ) const; \
|
||||||
|
virtual char as_char( long n ) const; \
|
||||||
|
virtual short as_short( long n ) const; \
|
||||||
|
virtual int as_int( long n ) const; \
|
||||||
|
virtual int as_nclong( long n ) const; \
|
||||||
|
virtual long as_long( long n ) const; \
|
||||||
|
virtual float as_float( long n ) const; \
|
||||||
|
virtual double as_double( long n ) const; \
|
||||||
|
virtual char* as_string( long n ) const; \
|
||||||
|
virtual int invalid( void ) const; \
|
||||||
|
private: \
|
||||||
|
TYPE* the_values; \
|
||||||
|
std::ostream& print(std::ostream&) const; \
|
||||||
|
};
|
||||||
|
|
||||||
|
#define NcTypeEnum(TYPE) makename2(_nc__,TYPE)
|
||||||
|
#define _nc__ncbyte ncByte
|
||||||
|
#define _nc__char ncChar
|
||||||
|
#define _nc__short ncShort
|
||||||
|
#define _nc__int ncInt
|
||||||
|
#define _nc__nclong ncLong
|
||||||
|
#define _nc__long ncLong
|
||||||
|
#define _nc__float ncFloat
|
||||||
|
#define _nc__double ncDouble
|
||||||
|
#define NcValuesimplement(TYPE) \
|
||||||
|
NcVal(TYPE)::NcVal(TYPE)( void ) \
|
||||||
|
: NcValues(NcTypeEnum(TYPE), 0), the_values(0) \
|
||||||
|
{} \
|
||||||
|
\
|
||||||
|
NcVal(TYPE)::NcVal(TYPE)(long num, const TYPE* vals) \
|
||||||
|
: NcValues(NcTypeEnum(TYPE), num) \
|
||||||
|
{ \
|
||||||
|
the_values = new TYPE[num]; \
|
||||||
|
for(int i = 0; i < num; i++) \
|
||||||
|
the_values[i] = vals[i]; \
|
||||||
|
} \
|
||||||
|
\
|
||||||
|
NcVal(TYPE)::NcVal(TYPE)(long num) \
|
||||||
|
: NcValues(NcTypeEnum(TYPE), num), the_values(new TYPE[num]) \
|
||||||
|
{} \
|
||||||
|
\
|
||||||
|
NcVal(TYPE)::NcVal(TYPE)(const NcVal(TYPE)& v) : \
|
||||||
|
NcValues(v) \
|
||||||
|
{ \
|
||||||
|
delete[] the_values; \
|
||||||
|
the_values = new TYPE[v.the_number]; \
|
||||||
|
for(int i = 0; i < v.the_number; i++) \
|
||||||
|
the_values[i] = v.the_values[i]; \
|
||||||
|
} \
|
||||||
|
\
|
||||||
|
NcVal(TYPE)& NcVal(TYPE)::operator=(const NcVal(TYPE)& v) \
|
||||||
|
{ \
|
||||||
|
if ( &v != this) { \
|
||||||
|
NcValues::operator=(v); \
|
||||||
|
delete[] the_values; \
|
||||||
|
the_values = new TYPE[v.the_number]; \
|
||||||
|
for(int i = 0; i < v.the_number; i++) \
|
||||||
|
the_values[i] = v.the_values[i]; \
|
||||||
|
} \
|
||||||
|
return *this; \
|
||||||
|
} \
|
||||||
|
\
|
||||||
|
void* NcVal(TYPE)::base( void ) const \
|
||||||
|
{ \
|
||||||
|
return the_values; \
|
||||||
|
} \
|
||||||
|
\
|
||||||
|
NcVal(TYPE)::~NcVal(TYPE)( void ) \
|
||||||
|
{ \
|
||||||
|
delete[] the_values; \
|
||||||
|
} \
|
||||||
|
\
|
||||||
|
int NcVal(TYPE)::invalid( void ) const \
|
||||||
|
{ \
|
||||||
|
for(int i=0;i<the_number;i++) \
|
||||||
|
if (the_values[i] == makename2(ncBad_,TYPE)) return 1; \
|
||||||
|
return 0; \
|
||||||
|
} \
|
||||||
|
|
||||||
|
|
||||||
|
#define Ncbytes_for_one_implement(TYPE) \
|
||||||
|
int NcVal(TYPE)::bytes_for_one( void ) const \
|
||||||
|
{ \
|
||||||
|
return sizeof(TYPE); \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define as_ncbyte_implement(TYPE) \
|
||||||
|
ncbyte NcVal(TYPE)::as_ncbyte( long n ) const \
|
||||||
|
{ \
|
||||||
|
if (the_values[n] < 0 || the_values[n] > UCHAR_MAX) \
|
||||||
|
return ncBad_byte; \
|
||||||
|
return (ncbyte) the_values[n]; \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define as_char_implement(TYPE) \
|
||||||
|
char NcVal(TYPE)::as_char( long n ) const \
|
||||||
|
{ \
|
||||||
|
if (the_values[n] < CHAR_MIN || the_values[n] > CHAR_MAX) \
|
||||||
|
return ncBad_char; \
|
||||||
|
return (char) the_values[n]; \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define as_short_implement(TYPE) \
|
||||||
|
short NcVal(TYPE)::as_short( long n ) const \
|
||||||
|
{ \
|
||||||
|
if (the_values[n] < SHRT_MIN || the_values[n] > SHRT_MAX) \
|
||||||
|
return ncBad_short; \
|
||||||
|
return (short) the_values[n]; \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define NCINT_MIN INT_MIN
|
||||||
|
#define NCINT_MAX INT_MAX
|
||||||
|
#define as_int_implement(TYPE) \
|
||||||
|
int NcVal(TYPE)::as_int( long n ) const \
|
||||||
|
{ \
|
||||||
|
if (the_values[n] < NCINT_MIN || the_values[n] > NCINT_MAX) \
|
||||||
|
return ncBad_int; \
|
||||||
|
return (int) the_values[n]; \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define NCLONG_MIN INT_MIN
|
||||||
|
#define NCLONG_MAX INT_MAX
|
||||||
|
#define as_nclong_implement(TYPE) \
|
||||||
|
nclong NcVal(TYPE)::as_nclong( long n ) const \
|
||||||
|
{ \
|
||||||
|
if (the_values[n] < NCLONG_MIN || the_values[n] > NCLONG_MAX) \
|
||||||
|
return ncBad_nclong; \
|
||||||
|
return (nclong) the_values[n]; \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define as_long_implement(TYPE) \
|
||||||
|
long NcVal(TYPE)::as_long( long n ) const \
|
||||||
|
{ \
|
||||||
|
if (the_values[n] < LONG_MIN || the_values[n] > LONG_MAX) \
|
||||||
|
return ncBad_long; \
|
||||||
|
return (long) the_values[n]; \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define as_float_implement(TYPE) \
|
||||||
|
inline float NcVal(TYPE)::as_float( long n ) const \
|
||||||
|
{ \
|
||||||
|
return (float) the_values[n]; \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define as_double_implement(TYPE) \
|
||||||
|
inline double NcVal(TYPE)::as_double( long n ) const \
|
||||||
|
{ \
|
||||||
|
return (double) the_values[n]; \
|
||||||
|
}
|
||||||
|
|
||||||
|
#define as_string_implement(TYPE) \
|
||||||
|
char* NcVal(TYPE)::as_string( long n ) const \
|
||||||
|
{ \
|
||||||
|
char* s = new char[32]; \
|
||||||
|
std::ostringstream ostr; \
|
||||||
|
ostr << the_values[n]; \
|
||||||
|
ostr.str().copy(s, std::string::npos); \
|
||||||
|
s[ostr.str().length()] = 0; \
|
||||||
|
return s; \
|
||||||
|
}
|
||||||
|
|
||||||
|
class NcValues // ABC for value blocks
|
||||||
|
{
|
||||||
|
public:
|
||||||
|
NcValues( void );
|
||||||
|
NcValues(NcType, long);
|
||||||
|
virtual ~NcValues( void );
|
||||||
|
virtual long num( void );
|
||||||
|
virtual std::ostream& print(std::ostream&) const = 0;
|
||||||
|
virtual void* base( void ) const = 0;
|
||||||
|
virtual int bytes_for_one( void ) const = 0;
|
||||||
|
|
||||||
|
// The following member functions provide conversions from the value
|
||||||
|
// type to a desired basic type. If the value is out of range, the
|
||||||
|
// default "fill-value" for the appropriate type is returned.
|
||||||
|
virtual ncbyte as_ncbyte( long n ) const = 0; // nth value as a byte
|
||||||
|
virtual char as_char( long n ) const = 0; // nth value as char
|
||||||
|
virtual short as_short( long n ) const = 0; // nth value as short
|
||||||
|
virtual int as_int( long n ) const = 0; // nth value as int
|
||||||
|
virtual int as_nclong( long n ) const = 0; // nth value as nclong
|
||||||
|
virtual long as_long( long n ) const = 0; // nth value as long
|
||||||
|
virtual float as_float( long n ) const = 0; // nth value as floating-point
|
||||||
|
virtual double as_double( long n ) const = 0; // nth value as double
|
||||||
|
virtual char* as_string( long n ) const = 0; // value as string
|
||||||
|
|
||||||
|
protected:
|
||||||
|
NcType the_type;
|
||||||
|
long the_number;
|
||||||
|
friend std::ostream& operator<< (std::ostream&, const NcValues&);
|
||||||
|
};
|
||||||
|
|
||||||
|
declare(NcValues,ncbyte)
|
||||||
|
declare(NcValues,char)
|
||||||
|
declare(NcValues,short)
|
||||||
|
declare(NcValues,int)
|
||||||
|
declare(NcValues,nclong)
|
||||||
|
declare(NcValues,long)
|
||||||
|
declare(NcValues,float)
|
||||||
|
declare(NcValues,double)
|
||||||
|
|
||||||
|
#endif
|
||||||
1676
dep/netcdfcxx_legacy/src/lib/netcdf.cpp
Normal file
92
dep/netcdfcxx_legacy/src/lib/netcdf_config.h
Normal file
@ -0,0 +1,92 @@
|
|||||||
|
/* config.h.in. Generated from configure.ac by autoheader. */
|
||||||
|
|
||||||
|
/* if true, run extra tests which may not work yet */
|
||||||
|
#undef EXTRA_TESTS
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <dlfcn.h> header file. */
|
||||||
|
#undef HAVE_DLFCN_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <inttypes.h> header file. */
|
||||||
|
#undef HAVE_INTTYPES_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <memory.h> header file. */
|
||||||
|
#undef HAVE_MEMORY_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the `nccreate' function. */
|
||||||
|
#undef HAVE_NCCREATE
|
||||||
|
|
||||||
|
/* Define to 1 if you have the `nc_def_opaque' function. */
|
||||||
|
#undef HAVE_NC_DEF_OPAQUE
|
||||||
|
|
||||||
|
/* Define to 1 if you have the `nc_set_log_level' function. */
|
||||||
|
#undef HAVE_NC_SET_LOG_LEVEL
|
||||||
|
|
||||||
|
/* Define to 1 if you have the `nc_use_parallel_enabled' function. */
|
||||||
|
#undef HAVE_NC_USE_PARALLEL_ENABLED
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <netcdf.h> header file. */
|
||||||
|
#undef HAVE_NETCDF_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <stdint.h> header file. */
|
||||||
|
#undef HAVE_STDINT_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <stdlib.h> header file. */
|
||||||
|
#define HAVE_STDLIB_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <strings.h> header file. */
|
||||||
|
#undef HAVE_STRINGS_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <string.h> header file. */
|
||||||
|
#undef HAVE_STRING_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <sys/stat.h> header file. */
|
||||||
|
#undef HAVE_SYS_STAT_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <sys/types.h> header file. */
|
||||||
|
#undef HAVE_SYS_TYPES_H
|
||||||
|
|
||||||
|
/* Define to 1 if you have the <unistd.h> header file. */
|
||||||
|
#undef HAVE_UNISTD_H
|
||||||
|
|
||||||
|
/* do large file tests */
|
||||||
|
#undef LARGE_FILE_TESTS
|
||||||
|
|
||||||
|
/* Define to the sub-directory in which libtool stores uninstalled libraries.
|
||||||
|
*/
|
||||||
|
#undef LT_OBJDIR
|
||||||
|
|
||||||
|
/* Name of package */
|
||||||
|
#undef PACKAGE
|
||||||
|
|
||||||
|
/* Define to the address where bug reports for this package should be sent. */
|
||||||
|
#undef PACKAGE_BUGREPORT
|
||||||
|
|
||||||
|
/* Define to the full name of this package. */
|
||||||
|
#undef PACKAGE_NAME
|
||||||
|
|
||||||
|
/* Define to the full name and version of this package. */
|
||||||
|
#undef PACKAGE_STRING
|
||||||
|
|
||||||
|
/* Define to the one symbol short name of this package. */
|
||||||
|
#undef PACKAGE_TARNAME
|
||||||
|
|
||||||
|
/* Define to the home page for this package. */
|
||||||
|
#undef PACKAGE_URL
|
||||||
|
|
||||||
|
/* Define to the version of this package. */
|
||||||
|
#undef PACKAGE_VERSION
|
||||||
|
|
||||||
|
/* Define to 1 if you have the ANSI C header files. */
|
||||||
|
#undef STDC_HEADERS
|
||||||
|
|
||||||
|
/* Place to put very large netCDF test files. */
|
||||||
|
#undef TEMP_LARGE
|
||||||
|
|
||||||
|
/* Version number of package */
|
||||||
|
#undef VERSION
|
||||||
|
|
||||||
|
/* Number of bits in a file offset, on hosts where this is settable. */
|
||||||
|
#undef _FILE_OFFSET_BITS
|
||||||
|
|
||||||
|
/* Define for large files, on AIX-style hosts. */
|
||||||
|
#undef _LARGE_FILES
|
||||||
469
dep/netcdfcxx_legacy/src/lib/netcdfcpp.h
Normal file
@ -0,0 +1,469 @@
|
|||||||
|
/*********************************************************************
|
||||||
|
* Copyright 1992, University Corporation for Atmospheric Research
|
||||||
|
* See netcdf/README file for copying and redistribution conditions.
|
||||||
|
*
|
||||||
|
* Purpose: C++ class interface for netCDF
|
||||||
|
*
|
||||||
|
* $Header: /upc/share/CVS/netcdf-3/cxx/netcdfcpp.h,v 1.15 2009/03/10 15:20:54 russ Exp $
|
||||||
|
*********************************************************************/
|
||||||
|
|
||||||
|
#ifndef NETCDF_HH
|
||||||
|
#define NETCDF_HH
|
||||||
|
|
||||||
|
#include "ncvalues.h" // arrays that know their element type
|
||||||
|
|
||||||
|
typedef const char* NcToken; // names for netCDF objects
|
||||||
|
typedef unsigned int NcBool; // many members return 0 on failure
|
||||||
|
|
||||||
|
class NcDim; // dimensions
|
||||||
|
class NcVar; // variables
|
||||||
|
class NcAtt; // attributes
|
||||||
|
|
||||||
|
/*
|
||||||
|
* ***********************************************************************
|
||||||
|
* A netCDF file.
|
||||||
|
* ***********************************************************************
|
||||||
|
*/
|
||||||
|
class NcFile
|
||||||
|
{
|
||||||
|
public:
|
||||||
|
|
||||||
|
virtual ~NcFile( void );
|
||||||
|
|
||||||
|
enum FileMode {
|
||||||
|
ReadOnly, // file exists, open read-only
|
||||||
|
Write, // file exists, open for writing
|
||||||
|
Replace, // create new file, even if already exists
|
||||||
|
New // create new file, fail if already exists
|
||||||
|
};
|
||||||
|
|
||||||
|
enum FileFormat {
|
||||||
|
Classic, // netCDF classic format (i.e. version 1 format)
|
||||||
|
Offset64Bits, // netCDF 64-bit offset format
|
||||||
|
Netcdf4, // netCDF-4 using HDF5 format
|
||||||
|
Netcdf4Classic, // netCDF-4 using HDF5 format using only netCDF-3 calls
|
||||||
|
BadFormat
|
||||||
|
};
|
||||||
|
|
||||||
|
NcFile( const char * path, FileMode = ReadOnly ,
|
||||||
|
size_t *bufrsizeptr = NULL, // optional tuning parameters
|
||||||
|
size_t initialsize = 0,
|
||||||
|
FileFormat = Classic );
|
||||||
|
|
||||||
|
NcBool is_valid( void ) const; // opened OK in ctr, still valid
|
||||||
|
|
||||||
|
int num_dims( void ) const; // number of dimensions
|
||||||
|
int num_vars( void ) const; // number of variables
|
||||||
|
int num_atts( void ) const; // number of (global) attributes
|
||||||
|
|
||||||
|
NcDim* get_dim( NcToken ) const; // dimension by name
|
||||||
|
NcVar* get_var( NcToken ) const; // variable by name
|
||||||
|
NcAtt* get_att( NcToken ) const; // global attribute by name
|
||||||
|
|
||||||
|
NcDim* get_dim( int ) const; // n-th dimension
|
||||||
|
NcVar* get_var( int ) const; // n-th variable
|
||||||
|
NcAtt* get_att( int ) const; // n-th global attribute
|
||||||
|
NcDim* rec_dim( void ) const; // unlimited dimension, if any
|
||||||
|
|
||||||
|
// Add new dimensions, variables, global attributes.
|
||||||
|
// These put the file in "define" mode, so could be expensive.
|
||||||
|
virtual NcDim* add_dim( NcToken dimname, long dimsize );
|
||||||
|
virtual NcDim* add_dim( NcToken dimname ); // unlimited
|
||||||
|
|
||||||
|
virtual NcVar* add_var( NcToken varname, NcType type, // scalar
|
||||||
|
const NcDim* dim0=0, // 1-dim
|
||||||
|
const NcDim* dim1=0, // 2-dim
|
||||||
|
const NcDim* dim2=0, // 3-dim
|
||||||
|
const NcDim* dim3=0, // 4-dim
|
||||||
|
const NcDim* dim4=0 ); // 5-dim
|
||||||
|
virtual NcVar* add_var( NcToken varname, NcType type, // n-dim
|
||||||
|
int ndims, const NcDim** dims );
|
||||||
|
|
||||||
|
NcBool add_att( NcToken attname, char ); // scalar attributes
|
||||||
|
NcBool add_att( NcToken attname, ncbyte );
|
||||||
|
NcBool add_att( NcToken attname, short );
|
||||||
|
NcBool add_att( NcToken attname, long );
|
||||||
|
NcBool add_att( NcToken attname, int );
|
||||||
|
NcBool add_att( NcToken attname, float );
|
||||||
|
NcBool add_att( NcToken attname, double );
|
||||||
|
NcBool add_att( NcToken attname, const char*); // string attribute
|
||||||
|
NcBool add_att( NcToken attname, int, const char* ); // vector attributes
|
||||||
|
NcBool add_att( NcToken attname, int, const ncbyte* );
|
||||||
|
NcBool add_att( NcToken attname, int, const short* );
|
||||||
|
NcBool add_att( NcToken attname, int, const long* );
|
||||||
|
NcBool add_att( NcToken attname, int, const int* );
|
||||||
|
NcBool add_att( NcToken attname, int, const float* );
|
||||||
|
NcBool add_att( NcToken attname, int, const double* );
|
||||||
|
|
||||||
|
enum FillMode {
|
||||||
|
Fill = NC_FILL, // prefill (default)
|
||||||
|
NoFill = NC_NOFILL, // don't prefill
|
||||||
|
Bad
|
||||||
|
};
|
||||||
|
|
||||||
|
NcBool set_fill( FillMode = Fill ); // set fill-mode
|
||||||
|
FillMode get_fill( void ) const; // get fill-mode
|
||||||
|
FileFormat get_format( void ) const; // get format version
|
||||||
|
|
||||||
|
NcBool sync( void ); // synchronize to disk
|
||||||
|
NcBool close( void ); // to close earlier than dtr
|
||||||
|
NcBool abort( void ); // back out of bad defines
|
||||||
|
|
||||||
|
// Needed by other Nc classes, but users will not need them
|
||||||
|
NcBool define_mode( void ); // leaves in define mode, if possible
|
||||||
|
NcBool data_mode( void ); // leaves in data mode, if possible
|
||||||
|
int id( void ) const; // id used by C interface
|
||||||
|
|
||||||
|
protected:
|
||||||
|
int the_id;
|
||||||
|
int in_define_mode;
|
||||||
|
FillMode the_fill_mode;
|
||||||
|
NcDim** dimensions;
|
||||||
|
NcVar** variables;
|
||||||
|
NcVar* globalv; // "variable" for global attributes
|
||||||
|
};
|
||||||
|
|
||||||
|
/*
|
||||||
|
* For backward compatibility. We used to derive NcOldFile and NcNewFile
|
||||||
|
* from NcFile, but that was over-zealous inheritance.
|
||||||
|
*/
|
||||||
|
#define NcOldFile NcFile
|
||||||
|
#define NcNewFile NcFile
|
||||||
|
#define Clobber Replace
|
||||||
|
#define NoClobber New
|
||||||
|
|
||||||
|
/*
|
||||||
|
* **********************************************************************
|
||||||
|
* A netCDF dimension, with a name and a size. These are only created
|
||||||
|
* by NcFile member functions, because they cannot exist independently
|
||||||
|
* of an open netCDF file.
|
||||||
|
* **********************************************************************
|
||||||
|
*/
|
||||||
|
class NcDim
|
||||||
|
{
|
||||||
|
public:
|
||||||
|
NcToken name( void ) const;
|
||||||
|
long size( void ) const;
|
||||||
|
NcBool is_valid( void ) const;
|
||||||
|
NcBool is_unlimited( void ) const;
|
||||||
|
NcBool rename( NcToken newname );
|
||||||
|
int id( void ) const;
|
||||||
|
NcBool sync( void );
|
||||||
|
|
||||||
|
private:
|
||||||
|
NcFile *the_file; // not const because of rename
|
||||||
|
int the_id;
|
||||||
|
char *the_name;
|
||||||
|
|
||||||
|
NcDim(NcFile*, int num); // existing dimension
|
||||||
|
NcDim(NcFile*, NcToken name, long sz); // defines a new dim
|
||||||
|
virtual ~NcDim( void );
|
||||||
|
|
||||||
|
// to construct dimensions, since constructor is private
|
||||||
|
friend class NcFile;
|
||||||
|
};
|
||||||
|
|
||||||
|
|
||||||
|
/*
|
||||||
|
* **********************************************************************
|
||||||
|
* Abstract base class for a netCDF variable or attribute, both of which
|
||||||
|
* have a name, a type, and associated values. These only exist as
|
||||||
|
* components of an open netCDF file.
|
||||||
|
* **********************************************************************
|
||||||
|
*/
|
||||||
|
class NcTypedComponent
|
||||||
|
{
|
||||||
|
public:
|
||||||
|
virtual ~NcTypedComponent( void ) {}
|
||||||
|
virtual NcToken name( void ) const = 0;
|
||||||
|
virtual NcType type( void ) const = 0;
|
||||||
|
virtual NcBool is_valid( void ) const = 0;
|
||||||
|
virtual long num_vals( void ) const = 0;
|
||||||
|
virtual NcBool rename( NcToken newname ) = 0;
|
||||||
|
virtual NcValues* values( void ) const = 0; // block of all values
|
||||||
|
|
||||||
|
// The following member functions provide conversions from the value
|
||||||
|
// type to a desired basic type. If the value is out of range,
|
||||||
|
// the default "fill-value" for the appropriate type is returned.
|
||||||
|
|
||||||
|
virtual ncbyte as_ncbyte( long n ) const; // nth value as an unsgnd char
|
||||||
|
virtual char as_char( long n ) const; // nth value as char
|
||||||
|
virtual short as_short( long n ) const; // nth value as short
|
||||||
|
virtual int as_int( long n ) const; // nth value as int
|
||||||
|
virtual int as_nclong( long n ) const; // nth value as nclong (deprecated)
|
||||||
|
virtual long as_long( long n ) const; // nth value as long
|
||||||
|
virtual float as_float( long n ) const; // nth value as floating-point
|
||||||
|
virtual double as_double( long n ) const; // nth value as double
|
||||||
|
virtual char* as_string( long n ) const; // nth value as string
|
||||||
|
|
||||||
|
protected:
|
||||||
|
NcFile *the_file;
|
||||||
|
NcTypedComponent( NcFile* );
|
||||||
|
virtual NcValues* get_space( long numVals = 0 ) const; // to hold values
|
||||||
|
};
|
||||||
|
|
||||||
|
|
||||||
|
/*
|
||||||
|
* **********************************************************************
|
||||||
|
* netCDF variables. In addition to a name and a type, these also have
|
||||||
|
* a shape, given by a list of dimensions
|
||||||
|
* **********************************************************************
|
||||||
|
*/
|
||||||
|
class NcVar : public NcTypedComponent
|
||||||
|
{
|
||||||
|
public:
|
||||||
|
virtual ~NcVar( void );
|
||||||
|
NcToken name( void ) const;
|
||||||
|
NcType type( void ) const;
|
||||||
|
NcBool is_valid( void ) const;
|
||||||
|
int num_dims( void ) const; // dimensionality of variable
|
||||||
|
NcDim* get_dim( int ) const; // n-th dimension
|
||||||
|
long* edges( void ) const; // dimension sizes
|
||||||
|
int num_atts( void ) const; // number of attributes
|
||||||
|
NcAtt* get_att( NcToken ) const; // attribute by name
|
||||||
|
NcAtt* get_att( int ) const; // n-th attribute
|
||||||
|
long num_vals( void ) const; // product of dimension sizes
|
||||||
|
NcValues* values( void ) const; // all values
|
||||||
|
|
||||||
|
// Put scalar or 1, ..., 5 dimensional arrays by providing enough
|
||||||
|
// arguments. Arguments are edge lengths, and their number must not
|
||||||
|
// exceed variable's dimensionality. Start corner is [0,0,..., 0] by
|
||||||
|
// default, but may be reset using the set_cur() member. FALSE is
|
||||||
|
// returned if type of values does not match type for variable.
|
||||||
|
NcBool put( const ncbyte* vals,
|
||||||
|
long c0=0, long c1=0, long c2=0, long c3=0, long c4=0 );
|
||||||
|
NcBool put( const char* vals,
|
||||||
|
long c0=0, long c1=0, long c2=0, long c3=0, long c4=0 );
|
||||||
|
NcBool put( const short* vals,
|
||||||
|
long c0=0, long c1=0, long c2=0, long c3=0, long c4=0 );
|
||||||
|
NcBool put( const int* vals,
|
||||||
|
long c0=0, long c1=0, long c2=0, long c3=0, long c4=0 );
|
||||||
|
NcBool put( const long* vals,
|
||||||
|
long c0=0, long c1=0, long c2=0, long c3=0, long c4=0 );
|
||||||
|
NcBool put( const float* vals,
|
||||||
|
long c0=0, long c1=0, long c2=0, long c3=0, long c4=0 );
|
||||||
|
NcBool put( const double* vals,
|
||||||
|
long c0=0, long c1=0, long c2=0, long c3=0, long c4=0 );
|
||||||
|
|
||||||
|
// Put n-dimensional arrays, starting at [0, 0, ..., 0] by default,
|
||||||
|
// may be reset with set_cur().
|
||||||
|
NcBool put( const ncbyte* vals, const long* counts );
|
||||||
|
NcBool put( const char* vals, const long* counts );
|
||||||
|
NcBool put( const short* vals, const long* counts );
|
||||||
|
NcBool put( const int* vals, const long* counts );
|
||||||
|
NcBool put( const long* vals, const long* counts );
|
||||||
|
NcBool put( const float* vals, const long* counts );
|
||||||
|
NcBool put( const double* vals, const long* counts );
|
||||||
|
|
||||||
|
// Get scalar or 1, ..., 5 dimensional arrays by providing enough
|
||||||
|
// arguments. Arguments are edge lengths, and their number must not
|
||||||
|
// exceed variable's dimensionality. Start corner is [0,0,..., 0] by
|
||||||
|
// default, but may be reset using the set_cur() member.
|
||||||
|
NcBool get( ncbyte* vals, long c0=0, long c1=0,
|
||||||
|
long c2=0, long c3=0, long c4=0 ) const;
|
||||||
|
NcBool get( char* vals, long c0=0, long c1=0,
|
||||||
|
long c2=0, long c3=0, long c4=0 ) const;
|
||||||
|
NcBool get( short* vals, long c0=0, long c1=0,
|
||||||
|
long c2=0, long c3=0, long c4=0 ) const;
|
||||||
|
NcBool get( int* vals, long c0=0, long c1=0,
|
||||||
|
long c2=0, long c3=0, long c4=0 ) const;
|
||||||
|
NcBool get( long* vals, long c0=0, long c1=0,
|
||||||
|
long c2=0, long c3=0, long c4=0 ) const;
|
||||||
|
NcBool get( float* vals, long c0=0, long c1=0,
|
||||||
|
long c2=0, long c3=0, long c4=0 ) const;
|
||||||
|
NcBool get( double* vals, long c0=0, long c1=0,
|
||||||
|
long c2=0, long c3=0, long c4=0 ) const;
|
||||||
|
|
||||||
|
// Get n-dimensional arrays, starting at [0, 0, ..., 0] by default,
|
||||||
|
// may be reset with set_cur().
|
||||||
|
NcBool get( ncbyte* vals, const long* counts ) const;
|
||||||
|
NcBool get( char* vals, const long* counts ) const;
|
||||||
|
NcBool get( short* vals, const long* counts ) const;
|
||||||
|
NcBool get( int* vals, const long* counts ) const;
|
||||||
|
NcBool get( long* vals, const long* counts ) const;
|
||||||
|
NcBool get( float* vals, const long* counts ) const;
|
||||||
|
NcBool get( double* vals, const long* counts ) const;
|
||||||
|
|
||||||
|
NcBool set_cur(long c0=-1, long c1=-1, long c2=-1,
|
||||||
|
long c3=-1, long c4=-1);
|
||||||
|
NcBool set_cur(long* cur);
|
||||||
|
|
||||||
|
// these put file in define mode, so could be expensive
|
||||||
|
NcBool add_att( NcToken, char ); // add scalar attributes
|
||||||
|
NcBool add_att( NcToken, ncbyte );
|
||||||
|
NcBool add_att( NcToken, short );
|
||||||
|
NcBool add_att( NcToken, int );
|
||||||
|
NcBool add_att( NcToken, long );
|
||||||
|
NcBool add_att( NcToken, float );
|
||||||
|
NcBool add_att( NcToken, double );
|
||||||
|
NcBool add_att( NcToken, const char* ); // string attribute
|
||||||
|
NcBool add_att( NcToken, int, const char* ); // vector attributes
|
||||||
|
NcBool add_att( NcToken, int, const ncbyte* );
|
||||||
|
NcBool add_att( NcToken, int, const short* );
|
||||||
|
NcBool add_att( NcToken, int, const int* );
|
||||||
|
NcBool add_att( NcToken, int, const long* );
|
||||||
|
NcBool add_att( NcToken, int, const float* );
|
||||||
|
NcBool add_att( NcToken, int, const double* );
|
||||||
|
|
||||||
|
NcBool rename( NcToken newname );
|
||||||
|
|
||||||
|
long rec_size ( void ); // number of values per record
|
||||||
|
long rec_size ( NcDim* ); // number of values per dimension slice
|
||||||
|
|
||||||
|
// Though following are intended for record variables, they also work
|
||||||
|
// for other variables, using first dimension as record dimension.
|
||||||
|
|
||||||
|
// Get a record's worth of data
|
||||||
|
NcValues *get_rec(void); // get current record
|
||||||
|
NcValues *get_rec(long rec); // get specified record
|
||||||
|
NcValues *get_rec(NcDim* d); // get current dimension slice
|
||||||
|
NcValues *get_rec(NcDim* d, long slice); // get specified dimension slice
|
||||||
|
|
||||||
|
// Put a record's worth of data in current record
|
||||||
|
NcBool put_rec( const ncbyte* vals );
|
||||||
|
NcBool put_rec( const char* vals );
|
||||||
|
NcBool put_rec( const short* vals );
|
||||||
|
NcBool put_rec( const int* vals );
|
||||||
|
NcBool put_rec( const long* vals );
|
||||||
|
NcBool put_rec( const float* vals );
|
||||||
|
NcBool put_rec( const double* vals );
|
||||||
|
|
||||||
|
// Put a dimension slice worth of data in current dimension slice
|
||||||
|
NcBool put_rec( NcDim* d, const ncbyte* vals );
|
||||||
|
NcBool put_rec( NcDim* d, const char* vals );
|
||||||
|
NcBool put_rec( NcDim* d, const short* vals );
|
||||||
|
NcBool put_rec( NcDim* d, const int* vals );
|
||||||
|
NcBool put_rec( NcDim* d, const long* vals );
|
||||||
|
NcBool put_rec( NcDim* d, const float* vals );
|
||||||
|
NcBool put_rec( NcDim* d, const double* vals );
|
||||||
|
|
||||||
|
// Put a record's worth of data in specified record
|
||||||
|
NcBool put_rec( const ncbyte* vals, long rec );
|
||||||
|
NcBool put_rec( const char* vals, long rec );
|
||||||
|
NcBool put_rec( const short* vals, long rec );
|
||||||
|
NcBool put_rec( const int* vals, long rec );
|
||||||
|
NcBool put_rec( const long* vals, long rec );
|
||||||
|
NcBool put_rec( const float* vals, long rec );
|
||||||
|
NcBool put_rec( const double* vals, long rec );
|
||||||
|
|
||||||
|
// Put a dimension slice worth of data in specified dimension slice
|
||||||
|
NcBool put_rec( NcDim* d, const ncbyte* vals, long slice );
|
||||||
|
NcBool put_rec( NcDim* d, const char* vals, long slice );
|
||||||
|
NcBool put_rec( NcDim* d, const short* vals, long slice );
|
||||||
|
NcBool put_rec( NcDim* d, const int* vals, long slice );
|
||||||
|
NcBool put_rec( NcDim* d, const long* vals, long slice );
|
||||||
|
NcBool put_rec( NcDim* d, const float* vals, long slice );
|
||||||
|
NcBool put_rec( NcDim* d, const double* vals, long slice );
|
||||||
|
|
||||||
|
// Get first record index corresponding to specified key value(s)
|
||||||
|
long get_index( const ncbyte* vals );
|
||||||
|
long get_index( const char* vals );
|
||||||
|
long get_index( const short* vals );
|
||||||
|
long get_index( const int* vals );
|
||||||
|
long get_index( const long* vals );
|
||||||
|
long get_index( const float* vals );
|
||||||
|
long get_index( const double* vals );
|
||||||
|
|
||||||
|
// Get first index of specified dimension corresponding to key values
|
||||||
|
long get_index( NcDim* d, const ncbyte* vals );
|
||||||
|
long get_index( NcDim* d, const char* vals );
|
||||||
|
long get_index( NcDim* d, const short* vals );
|
||||||
|
long get_index( NcDim* d, const int* vals );
|
||||||
|
long get_index( NcDim* d, const long* vals );
|
||||||
|
long get_index( NcDim* d, const float* vals );
|
||||||
|
long get_index( NcDim* d, const double* vals );
|
||||||
|
|
||||||
|
// Set current record
|
||||||
|
void set_rec ( long rec );
|
||||||
|
// Set current dimension slice
|
||||||
|
void set_rec ( NcDim* d, long slice );
|
||||||
|
|
||||||
|
int id( void ) const; // rarely needed, C interface id
|
||||||
|
NcBool sync( void );
|
||||||
|
|
||||||
|
private:
|
||||||
|
int dim_to_index(NcDim* rdim);
|
||||||
|
int the_id;
|
||||||
|
long* the_cur;
|
||||||
|
char* the_name;
|
||||||
|
long* cur_rec;
|
||||||
|
|
||||||
|
// private constructors because only an NcFile creates these
|
||||||
|
NcVar( void );
|
||||||
|
NcVar(NcFile*, int);
|
||||||
|
|
||||||
|
int attnum( NcToken attname ) const;
|
||||||
|
NcToken attname( int attnum ) const;
|
||||||
|
void init_cur( void );
|
||||||
|
|
||||||
|
// to make variables, since constructor is private
|
||||||
|
friend class NcFile;
|
||||||
|
};
|
||||||
|
|
||||||
|
|
||||||
|
/*
|
||||||
|
* **********************************************************************
|
||||||
|
* netCDF attributes. In addition to a name and a type, these are each
|
||||||
|
* associated with a specific variable, or are global to the file.
|
||||||
|
* **********************************************************************
|
||||||
|
*/
|
||||||
|
class NcAtt : public NcTypedComponent
|
||||||
|
{
|
||||||
|
public:
|
||||||
|
virtual ~NcAtt( void );
|
||||||
|
NcToken name( void ) const;
|
||||||
|
NcType type( void ) const;
|
||||||
|
NcBool is_valid( void ) const;
|
||||||
|
long num_vals( void ) const;
|
||||||
|
NcValues* values( void ) const;
|
||||||
|
NcBool rename( NcToken newname );
|
||||||
|
NcBool remove( void );
|
||||||
|
|
||||||
|
private:
|
||||||
|
const NcVar* the_variable;
|
||||||
|
char* the_name;
|
||||||
|
// protected constructors because only NcVars and NcFiles create
|
||||||
|
// attributes
|
||||||
|
NcAtt( NcFile*, const NcVar*, NcToken);
|
||||||
|
NcAtt( NcFile*, NcToken); // global attribute
|
||||||
|
|
||||||
|
// To make attributes, since constructor is private
|
||||||
|
friend class NcFile;
|
||||||
|
friend NcAtt* NcVar::get_att( NcToken ) const;
|
||||||
|
};
|
||||||
|
|
||||||
|
|
||||||
|
/*
|
||||||
|
* **********************************************************************
|
||||||
|
* To control error handling. Declaring an NcError object temporarily
|
||||||
|
* changes the error-handling behavior until the object is destroyed, at
|
||||||
|
* which time the previous error-handling behavior is restored.
|
||||||
|
* **********************************************************************
|
||||||
|
*/
|
||||||
|
class NcError {
|
||||||
|
public:
|
||||||
|
enum Behavior {
|
||||||
|
silent_nonfatal = 0,
|
||||||
|
silent_fatal = 1,
|
||||||
|
verbose_nonfatal = 2,
|
||||||
|
verbose_fatal = 3
|
||||||
|
};
|
||||||
|
|
||||||
|
// constructor saves previous error state, sets new state
|
||||||
|
NcError( Behavior b = verbose_fatal );
|
||||||
|
|
||||||
|
// destructor restores previous error state
|
||||||
|
virtual ~NcError( void );
|
||||||
|
|
||||||
|
int get_err( void ); // returns most recent error number
|
||||||
|
const char* get_errmsg( void ) {return nc_strerror(get_err());}
|
||||||
|
static int set_err( int err );
|
||||||
|
|
||||||
|
private:
|
||||||
|
int the_old_state;
|
||||||
|
int the_old_err;
|
||||||
|
static int ncopts;
|
||||||
|
static int ncerr;
|
||||||
|
};
|
||||||
|
|
||||||
|
#endif /* NETCDF_HH */
|
||||||
70
dep/tesseroids/CITATION.txt
Normal file
@ -0,0 +1,70 @@
|
|||||||
|
Citing
|
||||||
|
======
|
||||||
|
|
||||||
|
Geophysics paper
|
||||||
|
----------------
|
||||||
|
|
||||||
|
To cite *Tesseroids* in publications, please use our paper published in
|
||||||
|
*Geophysics*:
|
||||||
|
|
||||||
|
Uieda, L., V. Barbosa, and C. Braitenberg (2016), Tesseroids:
|
||||||
|
Forward-modeling gravitational fields in spherical coordinates, GEOPHYSICS,
|
||||||
|
F41-F48,
|
||||||
|
doi:`10.1190/geo2015-0204.1 <http://dx.doi.org/10.1190/geo2015-0204.1>`__.
|
||||||
|
|
||||||
|
You can download a copy of the `paper PDF
|
||||||
|
<http://www.leouieda.com/papers/paper-tesseroids-2016.html>`__ and see all
|
||||||
|
source code used in the paper at
|
||||||
|
`the Github repository <https://github.com/pinga-lab/paper-tesseroids>`__.
|
||||||
|
|
||||||
|
Please note that **citing the paper is prefered** over citing the previous
|
||||||
|
conference proceedings.
|
||||||
|
|
||||||
|
If you're a BibTeX user::
|
||||||
|
|
||||||
|
@article{uieda2016,
|
||||||
|
title = {Tesseroids: {{Forward}}-modeling gravitational fields in spherical coordinates},
|
||||||
|
author = {Uieda, L. and Barbosa, V. and Braitenberg, C.},
|
||||||
|
issn = {0016-8033},
|
||||||
|
doi = {10.1190/geo2015-0204.1},
|
||||||
|
url = {http://library.seg.org/doi/abs/10.1190/geo2015-0204.1},
|
||||||
|
journal = {GEOPHYSICS},
|
||||||
|
month = jul,
|
||||||
|
year = {2016},
|
||||||
|
pages = {F41--F48},
|
||||||
|
}
|
||||||
|
|
||||||
|
Source code
|
||||||
|
-----------
|
||||||
|
|
||||||
|
You can refer to individual versions of Tesseroids through their DOIs.
|
||||||
|
However, please **also cite the Geophysics paper**.
|
||||||
|
|
||||||
|
For example. if you want to mention that you used the 1.1.1 version,
|
||||||
|
you can go to :ref:`the Releases page <releases>` of the documentation
|
||||||
|
and get the DOI link for that version.
|
||||||
|
This link will not be broken, even if I move the site somewhere else.
|
||||||
|
|
||||||
|
You can also cite the specific version instead of just providing the link.
|
||||||
|
If you click of the DOI link for 1.1.1, the Zenodo page will
|
||||||
|
recommend that you cite it as:
|
||||||
|
|
||||||
|
Uieda, Leonardo. (2015). Tesseroids v1.1.1: Forward modeling of
|
||||||
|
gravitational fields in spherical coordinates. Zenodo. 10.5281/zenodo.15800
|
||||||
|
|
||||||
|
|
||||||
|
Conference proceeding
|
||||||
|
---------------------
|
||||||
|
|
||||||
|
The previous way citation for Tesseroids was a conference proceeding from the
|
||||||
|
2011 GOCE User Workshop:
|
||||||
|
|
||||||
|
Uieda, L., E. P. Bomfim, C. Braitenberg, and E. Molina (2011),
|
||||||
|
Optimal forward calculation method of the Marussi tensor
|
||||||
|
due to a geologic structure at GOCE height,
|
||||||
|
Proceedings of the 4th International GOCE User Workshop.
|
||||||
|
|
||||||
|
Download a `PDF version of the proceedings
|
||||||
|
<http://www.leouieda.com/pdf/goce-2011.pdf>`__.
|
||||||
|
You can also see the poster and source code at
|
||||||
|
the `Github repository <https://github.com/leouieda/goce2011>`__.
|
||||||
18
dep/tesseroids/CMakeLists.txt
Normal file
@ -0,0 +1,18 @@
|
|||||||
|
cmake_minimum_required(VERSION 3.15.2)
|
||||||
|
# 设置项目名称与语言
|
||||||
|
project(LibTess VERSION 1.6 LANGUAGES C)
|
||||||
|
# 添加配置配件编写的函数
|
||||||
|
include(CMakePackageConfigHelpers)
|
||||||
|
|
||||||
|
if(${CMAKE_HOST_SYSTEM_NAME} STREQUAL "Windows")
|
||||||
|
set(CMAKE_INSTALL_PREFIX D:/Library)
|
||||||
|
endif()
|
||||||
|
|
||||||
|
message(STATUS "Platform: " ${CMAKE_HOST_SYSTEM_NAME})
|
||||||
|
message(STATUS "Install prefix: " ${CMAKE_INSTALL_PREFIX})
|
||||||
|
message(STATUS "Build type: " ${CMAKE_BUILD_TYPE})
|
||||||
|
|
||||||
|
# 添加库源文件地址
|
||||||
|
add_subdirectory(lib)
|
||||||
|
add_subdirectory(toolkits)
|
||||||
|
add_subdirectory(test)
|
||||||
25
dep/tesseroids/LICENSE.txt
Normal file
@ -0,0 +1,25 @@
|
|||||||
|
Copyright (c) 2012-2017, Leonardo Uieda
|
||||||
|
All rights reserved.
|
||||||
|
|
||||||
|
Redistribution and use in source and binary forms, with or without modification,
|
||||||
|
are permitted provided that the following conditions are met:
|
||||||
|
|
||||||
|
* Redistributions of source code must retain the above copyright notice,
|
||||||
|
this list of conditions and the following disclaimer.
|
||||||
|
* Redistributions in binary form must reproduce the above copyright notice,
|
||||||
|
this list of conditions and the following disclaimer in the documentation
|
||||||
|
and/or other materials provided with the distribution.
|
||||||
|
* Neither the name of Leonardo Uieda nor the names of any contributors
|
||||||
|
may be used to endorse or promote products derived from this software
|
||||||
|
without specific prior written permission.
|
||||||
|
|
||||||
|
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND
|
||||||
|
ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED
|
||||||
|
WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
|
||||||
|
DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR
|
||||||
|
ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES
|
||||||
|
(INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES;
|
||||||
|
LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON
|
||||||
|
ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT
|
||||||
|
(INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS
|
||||||
|
SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
|
||||||
16
dep/tesseroids/LibTessConfig.cmake.in
Normal file
@ -0,0 +1,16 @@
|
|||||||
|
@PACKAGE_INIT@
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_VERSION "@PROJECT_VERSION@")
|
||||||
|
set_and_check(@PROJECT_NAME@_INSTALL_PREFIX "${PACKAGE_PREFIX_DIR}")
|
||||||
|
set_and_check(@PROJECT_NAME@_INC_DIR "${PACKAGE_PREFIX_DIR}/include")
|
||||||
|
set_and_check(@PROJECT_NAME@_INCULDE_DIR "${PACKAGE_PREFIX_DIR}/include")
|
||||||
|
set_and_check(@PROJECT_NAME@_LIB_DIR "${PACKAGE_PREFIX_DIR}/lib")
|
||||||
|
set_and_check(@PROJECT_NAME@_LIBRARY_DIR "${PACKAGE_PREFIX_DIR}/lib")
|
||||||
|
|
||||||
|
set(@PROJECT_NAME@_LIB tess)
|
||||||
|
set(@PROJECT_NAME@_LIBRARY tess)
|
||||||
|
|
||||||
|
# include target information
|
||||||
|
include("${CMAKE_CURRENT_LIST_DIR}/@PROJECT_NAME@Targets.cmake")
|
||||||
|
|
||||||
|
check_required_components(@PROJECT_NAME@)
|
||||||
210
dep/tesseroids/README.md
Normal file
@ -0,0 +1,210 @@
|
|||||||
|
# 
|
||||||
|
|
||||||
|
[Documentation](http://tesseroids.leouieda.com) |
|
||||||
|
[Download](https://github.com/leouieda/tesseroids/releases)
|
||||||
|
|
||||||
|
[](https://github.com/leouieda/tesseroids/releases)
|
||||||
|
[](https://travis-ci.org/leouieda/tesseroids)
|
||||||
|
[](https://github.com/leouieda/tesseroids/blob/master/LICENSE.txt)
|
||||||
|
[](http://dx.doi.org/10.5281/zenodo.582366)
|
||||||
|
|
||||||
|
|
||||||
|
*Forward modeling of gravitational fields in spherical coordinates.*
|
||||||
|
|
||||||
|
Developed by [Leonardo Uieda](http://www.leouieda.com)
|
||||||
|
in cooperation with [Carla Braitenberg](http://lithoflex.org/).
|
||||||
|
|
||||||
|
## About
|
||||||
|
|
||||||
|
*Tesseroids* is a collection of **command-line tools**
|
||||||
|
for modeling the gravitational potential, acceleration, and
|
||||||
|
gradient (Marussi) tensor.
|
||||||
|
|
||||||
|
The mass models can be made of right rectangular prisms or tesseroids
|
||||||
|
(spherical prisms).
|
||||||
|
Computation for rectangular prisms can be made in Cartesian or spherical
|
||||||
|
(geocentric) coordinates.
|
||||||
|
|
||||||
|
[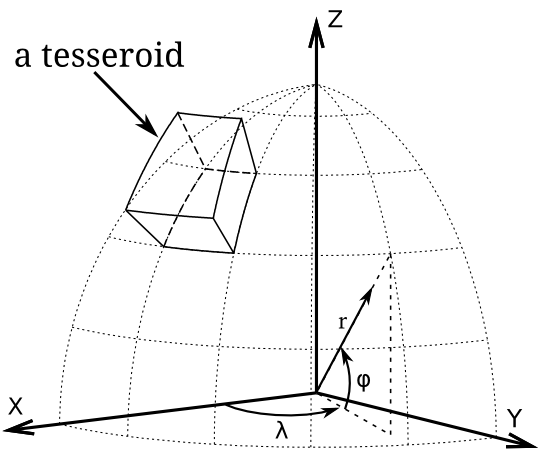](http://tesseroids.leouieda.com/en/latest/theory.html#what-is-a-tesseroid-anyway)
|
||||||
|
|
||||||
|
## License
|
||||||
|
|
||||||
|
*Tesseroids* is [free software](http://www.fsf.org/about/what-is-free-software)
|
||||||
|
made available under the terms of the
|
||||||
|
BSD 3-clause license.
|
||||||
|
See [LICENSE.txt](https://github.com/leouieda/tesseroids/blob/master/LICENSE.txt).
|
||||||
|
|
||||||
|
## Citing
|
||||||
|
|
||||||
|
*Tesseroids* is research software made by scientists.
|
||||||
|
If you use it in your research,
|
||||||
|
please **cite** our *Geophysics* paper in your publications:
|
||||||
|
|
||||||
|
> Uieda, L., V. Barbosa, and C. Braitenberg (2016), Tesseroids: Forward-modeling gravitational fields in spherical coordinates, GEOPHYSICS, F41-F48, doi:[10.1190/geo2015-0204.1](http://dx.doi.org/10.1190/geo2015-0204.1).
|
||||||
|
|
||||||
|
You can download a copy of the paper PDF at
|
||||||
|
[leouieda.com/papers/paper-tesseroids-2016.html](http://www.leouieda.com/papers/paper-tesseroids-2016.html)
|
||||||
|
and see all source code used in the paper at the Github repository
|
||||||
|
[pinga-lab/paper-tesseroids](https://github.com/pinga-lab/paper-tesseroids).
|
||||||
|
|
||||||
|
See [CITATION.txt](https://github.com/leouieda/tesseroids/blob/master/CITATION.txt)
|
||||||
|
or the [Citing](http://tesseroids.leouieda.com/en/latest/citation.html)
|
||||||
|
page of the documentation for more information.
|
||||||
|
|
||||||
|
## Installing
|
||||||
|
|
||||||
|
The easiest way to install is to download the latest compiled binary
|
||||||
|
distribution from:
|
||||||
|
|
||||||
|
https://github.com/leouieda/tesseroids/releases/latest
|
||||||
|
|
||||||
|
We offer binaries for Windows (32 and 64 bit)
|
||||||
|
and GNU/Linux (32 and 64 bit).
|
||||||
|
|
||||||
|
Once downloaded, simply unpack the archive in the desired directory.
|
||||||
|
The executables will be in the `bin` folder.
|
||||||
|
For easier access to the programs, consider
|
||||||
|
[adding the bin folder to your PATH environment
|
||||||
|
variable](http://www.computerhope.com/issues/ch000549.htm).
|
||||||
|
|
||||||
|
## Getting started
|
||||||
|
|
||||||
|
Take a look at the examples in the
|
||||||
|
[Cookbook](http://tesseroids.leouieda.com/en/latest/cookbook.html).
|
||||||
|
They contain scripts that run *Tesseroids* and some Python code to plot the
|
||||||
|
results.
|
||||||
|
|
||||||
|
The documentation contains sections on
|
||||||
|
[the theory and equations](http://tesseroids.leouieda.com/en/latest/theory.html)
|
||||||
|
and [usage instructions](http://tesseroids.leouieda.com/en/latest/usage.html).
|
||||||
|
|
||||||
|
Also, all programs accept the `-h` flag to print the instructions for using
|
||||||
|
that particular program. For example:
|
||||||
|
|
||||||
|
$ tessgrd -h
|
||||||
|
Usage: tessgrd [PARAMS] [OPTIONS]
|
||||||
|
|
||||||
|
Make a regular grid of points.
|
||||||
|
|
||||||
|
All units either SI or degrees!
|
||||||
|
|
||||||
|
Output:
|
||||||
|
Printed to standard output (stdout) in the format:
|
||||||
|
lon1 lat1 height
|
||||||
|
lon2 lat1 height
|
||||||
|
... ... ...
|
||||||
|
lonNLON lat1 height
|
||||||
|
lon1 lat2 height
|
||||||
|
... ... ...
|
||||||
|
... ... ...
|
||||||
|
lonNLON latNLAT height
|
||||||
|
|
||||||
|
* Comments about the provenance of the data are inserted into
|
||||||
|
the top of the output
|
||||||
|
|
||||||
|
Parameters:
|
||||||
|
-r W/E/S/N: Bounding region of the grid.
|
||||||
|
-b NLON/NLAT: Number of grid points in the
|
||||||
|
longitudinal and latitudinal directions.
|
||||||
|
-z HEIGHT: Height of the grid with respect to the
|
||||||
|
mean Earth radius.
|
||||||
|
-h Print instructions.
|
||||||
|
--version Print version and license information.
|
||||||
|
|
||||||
|
Options:
|
||||||
|
-v Enable verbose printing to stderr.
|
||||||
|
-lFILENAME Print log messages to file FILENAME.
|
||||||
|
|
||||||
|
Part of the Tesseroids package.
|
||||||
|
Project site: <http://fatiando.org/software/tesseroids>
|
||||||
|
Report bugs at: <http://code.google.com/p/tesseroids/issues/list>
|
||||||
|
|
||||||
|
|
||||||
|
## Getting help
|
||||||
|
|
||||||
|
Write an e-mail to [Leonardo Uieda](http://www.leouieda.com/),
|
||||||
|
or [tweet](https://twitter.com/leouieda),
|
||||||
|
or [Google Hangout](https://plus.google.com/+LeonardoUieda).
|
||||||
|
**Even better**, submit a bug report/feature request/question to the
|
||||||
|
[Github issue tracker](https://github.com/leouieda/tesseroids/issues).
|
||||||
|
|
||||||
|
## Compiling from source
|
||||||
|
|
||||||
|
If you want to build *Tesseroids* from source, you'll need:
|
||||||
|
|
||||||
|
* A C compiler (preferably [GCC](http://gcc.gnu.org))
|
||||||
|
* The build tool [SCons](http://www.scons.org/)
|
||||||
|
|
||||||
|
### Setting up SCons
|
||||||
|
|
||||||
|
Tesseroids uses the build tool SCons.
|
||||||
|
A `SConstruct` file (`Makefile` equivalent)
|
||||||
|
is used to define the compilation rules.
|
||||||
|
The advantage of SCons over Make is that it automatically detects your system
|
||||||
|
settings.
|
||||||
|
You will have to download and install SCons
|
||||||
|
in order to easily compile Tesseroids.
|
||||||
|
SCons is available for both GNU/Linux and Windows
|
||||||
|
so compiling should work the same on both platforms.
|
||||||
|
|
||||||
|
SCons requires that you have [Python](http://www.python.org) installed.
|
||||||
|
Follow the instructions in the [SCons website](http://www.scons.org/)
|
||||||
|
to install it.
|
||||||
|
Python is usually installed by default on most GNU/Linux systems.
|
||||||
|
|
||||||
|
Under Windows you will have to put SCons on
|
||||||
|
your `PATH` environment variable
|
||||||
|
in order to use it from the command line.
|
||||||
|
It is usually located in the `Scripts` directory of your Python installation.
|
||||||
|
|
||||||
|
On GNU/Linux, SCons will generally use
|
||||||
|
the GCC compiler to compile sources.
|
||||||
|
On Windows it will search for an existing compiler.
|
||||||
|
We recommend that you install GCC on Windows using
|
||||||
|
[MinGW](http://mingw.org/).
|
||||||
|
|
||||||
|
### Compiling
|
||||||
|
|
||||||
|
Download a source distribution and
|
||||||
|
unpack the archive anywhere you want
|
||||||
|
(e.g., `~/tesseroids` or `C:\tesseroids` or whatever).
|
||||||
|
To compile,
|
||||||
|
open a terminal (or `cmd.exe` on Windows)
|
||||||
|
and go to the directory where you unpacked (use the `cd` command).
|
||||||
|
Then, type the following and hit `Enter`:
|
||||||
|
|
||||||
|
scons
|
||||||
|
|
||||||
|
If everything goes well, the compiled executables will be placed on a `bin`
|
||||||
|
folder.
|
||||||
|
|
||||||
|
To clean up the build (delete all generated files), run:
|
||||||
|
|
||||||
|
scons -c
|
||||||
|
|
||||||
|
If you get any strange errors or the code doesn't compile for some reason,
|
||||||
|
please [submit a bug report](https://github.com/leouieda/tesseroids/issues).
|
||||||
|
Don't forget to copy the output of running `scons`.
|
||||||
|
|
||||||
|
### Testing the build
|
||||||
|
|
||||||
|
After the compilation,
|
||||||
|
a program called `tesstest`
|
||||||
|
will be placed in the directory where you unpacked the source.
|
||||||
|
This program runs the [unit tests](https://en.wikipedia.org/wiki/Unit_testing)
|
||||||
|
for *Tesseroids* (sources in the `test` directory).
|
||||||
|
|
||||||
|
To run the test suite, simply execute `tesstest` with no arguments:
|
||||||
|
|
||||||
|
tesstest
|
||||||
|
|
||||||
|
or on GNU/Linux:
|
||||||
|
|
||||||
|
./tesstest
|
||||||
|
|
||||||
|
A summary of all tests (pass or fail) will be printed on the screen.
|
||||||
|
If all tests pass,
|
||||||
|
the compilation probably went well.
|
||||||
|
If any test fail,
|
||||||
|
please [submit a bug report](https://github.com/leouieda/tesseroids/issues)
|
||||||
|
with the output of running `tesstest`.
|
||||||
10
dep/tesseroids/cookbook/custom_ratio/custom_ratio.bat
Executable file
@ -0,0 +1,10 @@
|
|||||||
|
:: Calculate effect of the model at a low height using difference distance-size
|
||||||
|
:: ratios for the recursive division of tesseroids.
|
||||||
|
:: WARNING: This is only an example. You should not use the -t option in
|
||||||
|
:: practice
|
||||||
|
|
||||||
|
tessgrd -r-3/3/-3/3 -b50/50 -z4e03 | ^
|
||||||
|
tessgzz model.txt -t0.0001 -lratio1.log | ^
|
||||||
|
tessgzz model.txt -t0.5 -lratio2.log | ^
|
||||||
|
tessgzz model.txt -t1 -lratio3.log | ^
|
||||||
|
tessgzz model.txt -v -lratio-default.log > output.txt
|
||||||
11
dep/tesseroids/cookbook/custom_ratio/custom_ratio.sh
Executable file
@ -0,0 +1,11 @@
|
|||||||
|
#!/bin/bash
|
||||||
|
|
||||||
|
# Calculate effect of the model at a low height using difference distance-size
|
||||||
|
# ratios for the recursive division of tesseroids.
|
||||||
|
# WARNING: This is only an example. You should not use the -t option in practice
|
||||||
|
|
||||||
|
tessgrd -r-3/3/-3/3 -b50/50 -z4e03 | \
|
||||||
|
tessgzz model.txt -t0.0001 -lratio1.log | \
|
||||||
|
tessgzz model.txt -t0.5 -lratio2.log | \
|
||||||
|
tessgzz model.txt -t1 -lratio3.log | \
|
||||||
|
tessgzz model.txt -v -lratio-default.log > output.txt
|
||||||
2
dep/tesseroids/cookbook/custom_ratio/model.txt
Executable file
@ -0,0 +1,2 @@
|
|||||||
|
# Test tesseroid model file
|
||||||
|
-1.5 1.5 -1.5 1.5 0 -5000 200
|
||||||
18
dep/tesseroids/cookbook/custom_ratio/plot.py
Executable file
@ -0,0 +1,18 @@
|
|||||||
|
"""
|
||||||
|
Plot the columns of the output files
|
||||||
|
"""
|
||||||
|
import sys
|
||||||
|
from matplotlib import pyplot as plt
|
||||||
|
import numpy as np
|
||||||
|
|
||||||
|
data = np.loadtxt(sys.argv[1], unpack=True)
|
||||||
|
shape = (int(sys.argv[2]), int(sys.argv[3]))
|
||||||
|
lon = np.reshape(data[0], shape)
|
||||||
|
lat = np.reshape(data[1], shape)
|
||||||
|
for i, value in enumerate(data[3:]):
|
||||||
|
value = np.reshape(value, shape)
|
||||||
|
plt.figure(figsize=(4, 3))
|
||||||
|
plt.title("Column %d" % (i + 4))
|
||||||
|
plt.contourf(lon, lat, value, 50)
|
||||||
|
plt.colorbar()
|
||||||
|
plt.savefig('column%d.png' % (i + 4))
|
||||||
22805
dep/tesseroids/cookbook/dem_brasil/dem.xyz
Executable file
22
dep/tesseroids/cookbook/dem_brasil/dem_brasil.bat
Executable file
@ -0,0 +1,22 @@
|
|||||||
|
|
||||||
|
|
||||||
|
:: First, insert the density information into
|
||||||
|
:: the DEM file using the Python script.
|
||||||
|
python dem_density.py dem.xyz > dem-dens.txt
|
||||||
|
|
||||||
|
:: Next, use the modified DEM with tessmodgen
|
||||||
|
:: to create a tesseroid model
|
||||||
|
tessmodgen -s0.166667/0.166667 -z0 -v < dem-dens.txt ^
|
||||||
|
> dem-tess.txt
|
||||||
|
|
||||||
|
:: Calculate the GGT on a regular grid at 250km
|
||||||
|
:: use the -l option to log the processes to files
|
||||||
|
:: (usefull to diagnose when things go wrong)
|
||||||
|
:: The output is dumped to dem-ggt.txt
|
||||||
|
tessgrd -r-60/-45/-30/-15 -b50/50 -z250e03 | ^
|
||||||
|
tessgxx dem-tess.txt -lgxx.log | ^
|
||||||
|
tessgxy dem-tess.txt -lgxy.log | ^
|
||||||
|
tessgxz dem-tess.txt -lgxz.log | ^
|
||||||
|
tessgyy dem-tess.txt -lgyy.log | ^
|
||||||
|
tessgyz dem-tess.txt -lgyz.log | ^
|
||||||
|
tessgzz dem-tess.txt -lgzz.log -v > dem-ggt.txt
|
||||||
22
dep/tesseroids/cookbook/dem_brasil/dem_brasil.sh
Executable file
@ -0,0 +1,22 @@
|
|||||||
|
#!/bin/bash
|
||||||
|
|
||||||
|
# First, insert the density information into
|
||||||
|
# the DEM file using the Python script.
|
||||||
|
python dem_density.py dem.xyz > dem-dens.txt
|
||||||
|
|
||||||
|
# Next, use the modified DEM with tessmodgen
|
||||||
|
# to create a tesseroid model
|
||||||
|
tessmodgen -s0.166667/0.166667 -z0 -v < dem-dens.txt \
|
||||||
|
> dem-tess.txt
|
||||||
|
|
||||||
|
# Calculate the GGT on a regular grid at 250km
|
||||||
|
# use the -l option to log the processes to files
|
||||||
|
# (usefull to diagnose when things go wrong)
|
||||||
|
# The output is dumped to dem-ggt.txt
|
||||||
|
tessgrd -r-60/-45/-30/-15 -b50/50 -z250e03 | \
|
||||||
|
tessgxx dem-tess.txt -lgxx.log | \
|
||||||
|
tessgxy dem-tess.txt -lgxy.log | \
|
||||||
|
tessgxz dem-tess.txt -lgxz.log | \
|
||||||
|
tessgyy dem-tess.txt -lgyy.log | \
|
||||||
|
tessgyz dem-tess.txt -lgyz.log | \
|
||||||
|
tessgzz dem-tess.txt -lgzz.log -v > dem-ggt.txt
|
||||||
13
dep/tesseroids/cookbook/dem_brasil/dem_density.py
Executable file
@ -0,0 +1,13 @@
|
|||||||
|
"""
|
||||||
|
Assign density values for the DEM points.
|
||||||
|
"""
|
||||||
|
import sys
|
||||||
|
import numpy
|
||||||
|
|
||||||
|
lons, lats, heights = numpy.loadtxt(sys.argv[1], unpack=True)
|
||||||
|
|
||||||
|
for i in xrange(len(heights)):
|
||||||
|
if heights[i] >=0:
|
||||||
|
print "%lf %lf %lf %lf" % (lons[i], lats[i], heights[i], 2670.0)
|
||||||
|
else:
|
||||||
|
print "%lf %lf %lf %lf" % (lons[i], lats[i], heights[i], 1670.0)
|
||||||
117
dep/tesseroids/cookbook/dem_brasil/plot.py
Executable file
@ -0,0 +1,117 @@
|
|||||||
|
# Make some nice plots of the DEM, the densities used and the calculated GGT
|
||||||
|
import numpy
|
||||||
|
from matplotlib import pyplot as plt
|
||||||
|
from mpl_toolkits.basemap import Basemap
|
||||||
|
|
||||||
|
# Plot the DEM and density maps
|
||||||
|
################################################################################
|
||||||
|
lons, lats, heights, dens = numpy.loadtxt('dem-dens.txt', unpack=True)
|
||||||
|
nlons = 151 # Number of points in the longitude direction
|
||||||
|
nlats = len(lats)/nlons
|
||||||
|
|
||||||
|
# Convert the lists to 2D grids
|
||||||
|
glons = numpy.reshape(lons, (nlats, nlons))
|
||||||
|
glats = numpy.reshape(lats, (nlats, nlons))
|
||||||
|
gheights = numpy.reshape(heights, (nlats, nlons))
|
||||||
|
gdens = numpy.reshape(dens, (nlats, nlons))
|
||||||
|
|
||||||
|
# Set up a Mercator projection
|
||||||
|
bm = Basemap(projection='merc',
|
||||||
|
llcrnrlon=lons[0], llcrnrlat=lats[-1],
|
||||||
|
urcrnrlon=lons[-1], urcrnrlat=lats[0],
|
||||||
|
lon_0=lons[nlons//2], lat_0=lats[len(lats)//2],
|
||||||
|
resolution='l',
|
||||||
|
area_thresh=10000)
|
||||||
|
glons, glats = bm(glons, glats)
|
||||||
|
|
||||||
|
# Plot the DEM first
|
||||||
|
print "Plotting DEM"
|
||||||
|
plt.figure()
|
||||||
|
bm.drawmeridians(numpy.arange(lons[0]+5., lons[-1], 5.),
|
||||||
|
labels=[0,0,0,1], fontsize=12, linewidth=0.5)
|
||||||
|
bm.drawparallels(numpy.arange(lats[-1]+5., lats[0], 5.),
|
||||||
|
labels=[1,0,0,0], fontsize=12, linewidth=0.5)
|
||||||
|
bm.drawcoastlines(linewidth=1)
|
||||||
|
bm.drawmapboundary()
|
||||||
|
bm.drawcountries(linewidth=0.8)
|
||||||
|
# Do the pseudocolor plot
|
||||||
|
cf = bm.pcolor(glons, glats, gheights, cmap=plt.cm.gist_earth, \
|
||||||
|
vmin=-1000, vmax=1000)
|
||||||
|
cb = plt.colorbar()
|
||||||
|
cb.set_label("Height [m]")
|
||||||
|
# Plot the calculation area used later
|
||||||
|
w = -60
|
||||||
|
e = -45
|
||||||
|
s = -30
|
||||||
|
n = -15
|
||||||
|
areax, areay = bm([w, w, e, e, w], \
|
||||||
|
[n, s, s, n, n])
|
||||||
|
bm.plot(areax, areay, '-r', label="Computation grid", linewidth=1.8)
|
||||||
|
plt.legend(shadow=True, loc='lower right', prop={'size':10})
|
||||||
|
# Save a png figure
|
||||||
|
plt.savefig('dem.png')
|
||||||
|
|
||||||
|
# Now plot the densities
|
||||||
|
print "Plotting density model"
|
||||||
|
plt.figure()
|
||||||
|
bm.drawmeridians(numpy.arange(lons[0]+5., lons[-1], 5.),
|
||||||
|
labels=[0,0,0,1], fontsize=12, linewidth=0.5)
|
||||||
|
bm.drawparallels(numpy.arange(lats[-1]+5., lats[0], 5.),
|
||||||
|
labels=[1,0,0,0], fontsize=12, linewidth=0.5)
|
||||||
|
bm.drawcoastlines(linewidth=1)
|
||||||
|
bm.drawmapboundary()
|
||||||
|
bm.drawcountries(linewidth=0.8)
|
||||||
|
# Do the pseudocolor plot
|
||||||
|
cf = bm.pcolor(glons, glats, gdens, cmap=plt.cm.jet)
|
||||||
|
cb = plt.colorbar()
|
||||||
|
cb.set_label(r"Density [$g.cm^{-3}$]")
|
||||||
|
# Save a png figure
|
||||||
|
plt.savefig('dem-dens.png')
|
||||||
|
|
||||||
|
# Plot the GGT
|
||||||
|
################################################################################
|
||||||
|
print "Plotting GGT"
|
||||||
|
data = numpy.loadtxt('dem-ggt.txt')
|
||||||
|
lons, lats, heights, gxx, gxy, gxz, gyy, gyz, gzz = data.T
|
||||||
|
nlons = 50 # Number of points in the longitude direction
|
||||||
|
nlats = len(lats)/nlons
|
||||||
|
|
||||||
|
# Convert the lists to 2D grids
|
||||||
|
glons = numpy.reshape(lons, (nlats, nlons))
|
||||||
|
glats = numpy.reshape(lats, (nlats, nlons))
|
||||||
|
|
||||||
|
# Set up a Mercator projection
|
||||||
|
bm = Basemap(projection='merc', \
|
||||||
|
llcrnrlon=lons[0], llcrnrlat=lats[0], \
|
||||||
|
urcrnrlon=lons[-1], urcrnrlat=lats[-1], \
|
||||||
|
lon_0=lons[nlons//2], lat_0=lats[len(lats)//2],
|
||||||
|
resolution='l', area_thresh=10000)
|
||||||
|
glons, glats = bm(glons, glats)
|
||||||
|
|
||||||
|
# Plot each component
|
||||||
|
fig = plt.figure(figsize=(14,9))
|
||||||
|
plt.subplots_adjust(wspace=0.35)
|
||||||
|
titles = [r"$g_{xx}$", r"$g_{xy}$", r"$g_{xz}$", r"$g_{yy}$", r"$g_{yz}$",
|
||||||
|
r"$g_{zz}$"]
|
||||||
|
fields = [gxx, gxy, gxz, gyy, gyz, gzz]
|
||||||
|
for i, args in enumerate(zip(fields, titles)):
|
||||||
|
field, title = args
|
||||||
|
ax = plt.subplot(2, 3, i + 1, aspect='equal')
|
||||||
|
plt.title(title, fontsize=18)
|
||||||
|
# Make it a 2D grid
|
||||||
|
gfield = numpy.reshape(field, (nlats, nlons))
|
||||||
|
# Plot the coastlines and etc
|
||||||
|
mer = bm.drawmeridians(numpy.arange(lons[0]+3, lons[-1]-3, 3),
|
||||||
|
labels=[0,0,0,1], fontsize=9, linewidth=0.5)
|
||||||
|
bm.drawparallels(numpy.arange(lats[0]+3, lats[-1]-3, 3),
|
||||||
|
labels=[1,0,0,0], fontsize=9, linewidth=0.5)
|
||||||
|
bm.drawcoastlines(linewidth=1)
|
||||||
|
bm.drawmapboundary()
|
||||||
|
bm.drawcountries(linewidth=1)
|
||||||
|
bm.drawstates(linewidth=0.2)
|
||||||
|
# Make a pseudocolor plot
|
||||||
|
cf = bm.pcolor(glons, glats, gfield, cmap=plt.cm.jet)
|
||||||
|
cb = plt.colorbar(orientation='vertical', format='%.2f', shrink=0.8)
|
||||||
|
cb.set_label(r"$E\"otv\"os$")
|
||||||
|
# Save a png figure
|
||||||
|
plt.savefig('dem-ggt.png')
|
||||||
BIN
dep/tesseroids/cookbook/dem_brasil/sample-dem-dens.png
Executable file
|
After Width: | Height: | Size: 150 KiB |
22801
dep/tesseroids/cookbook/dem_brasil/sample-dem-dens.txt
Executable file
BIN
dep/tesseroids/cookbook/dem_brasil/sample-dem-ggt.png
Executable file
|
After Width: | Height: | Size: 372 KiB |
22806
dep/tesseroids/cookbook/dem_brasil/sample-dem-tess.txt
Executable file
BIN
dep/tesseroids/cookbook/dem_brasil/sample-dem.png
Executable file
|
After Width: | Height: | Size: 260 KiB |
3
dep/tesseroids/cookbook/simple_prism/model.txt
Executable file
@ -0,0 +1,3 @@
|
|||||||
|
# Test prism model file
|
||||||
|
2000 5000 2000 15000 0 5000 1000
|
||||||
|
10000 18000 10000 18000 0 5000 -1000
|
||||||
25
dep/tesseroids/cookbook/simple_prism/plot.py
Executable file
@ -0,0 +1,25 @@
|
|||||||
|
"""
|
||||||
|
Plot the columns of the output files
|
||||||
|
"""
|
||||||
|
import sys
|
||||||
|
import pylab
|
||||||
|
|
||||||
|
data = pylab.loadtxt(sys.argv[1], unpack=True)
|
||||||
|
shape = (int(sys.argv[2]), int(sys.argv[3]))
|
||||||
|
lon = pylab.reshape(data[0], shape)
|
||||||
|
lat = pylab.reshape(data[1], shape)
|
||||||
|
xmin, xmax = lon.min(), lon.max()
|
||||||
|
ymin, ymax = lat.min(), lat.max()
|
||||||
|
for i, value in enumerate(data[3:]):
|
||||||
|
value = pylab.reshape(value, shape)
|
||||||
|
pylab.figure(figsize=(4, 3))
|
||||||
|
pylab.title("Column %d" % (i + 4))
|
||||||
|
pylab.axis('scaled')
|
||||||
|
pylab.pcolor(lon, lat, value)
|
||||||
|
pylab.colorbar()
|
||||||
|
pylab.contour(lon, lat, value, 12, color='k')
|
||||||
|
#pylab.xlabel("Longitude")
|
||||||
|
#pylab.ylabel("Latitude")
|
||||||
|
pylab.xlim(xmin, xmax)
|
||||||
|
pylab.ylim(ymin, ymax)
|
||||||
|
pylab.savefig('column%d.png' % (i + 4))
|
||||||
11
dep/tesseroids/cookbook/simple_prism/simple_prism.bat
Executable file
@ -0,0 +1,11 @@
|
|||||||
|
|
||||||
|
|
||||||
|
:: Generate a regular grid, pipe it to all the computation programs,
|
||||||
|
:: and write the result to output.txt
|
||||||
|
|
||||||
|
tessgrd -r0/20000/0/20000 -b50/50 -z1000 | ^
|
||||||
|
prismpot model.txt | ^
|
||||||
|
prismgx model.txt | prismgy model.txt | prismgz model.txt | ^
|
||||||
|
prismgxx model.txt | prismgxy model.txt | ^
|
||||||
|
prismgxz model.txt | prismgyy model.txt | ^
|
||||||
|
prismgyz model.txt | prismgzz model.txt > output.txt
|
||||||
BIN
dep/tesseroids/cookbook/simple_prism/simple_prism.png
Executable file
|
After Width: | Height: | Size: 585 KiB |
11
dep/tesseroids/cookbook/simple_prism/simple_prism.sh
Executable file
@ -0,0 +1,11 @@
|
|||||||
|
#!/bin/bash
|
||||||
|
|
||||||
|
# Generate a regular grid, pipe it to all the computation programs,
|
||||||
|
# and write the result to output.txt
|
||||||
|
|
||||||
|
tessgrd -r0/20000/0/20000 -b50/50 -z1000 | \
|
||||||
|
prismpot model.txt | \
|
||||||
|
prismgx model.txt | prismgy model.txt | prismgz model.txt | \
|
||||||
|
prismgxx model.txt | prismgxy model.txt | \
|
||||||
|
prismgxz model.txt | prismgyy model.txt | \
|
||||||
|
prismgyz model.txt | prismgzz model.txt > output.txt
|
||||||
3
dep/tesseroids/cookbook/simple_tess/model.txt
Executable file
@ -0,0 +1,3 @@
|
|||||||
|
# Test tesseroid model file
|
||||||
|
10 20 10 20 0 -50000 200
|
||||||
|
-20 -10 -20 -10 0 -30000 -500
|
||||||
29
dep/tesseroids/cookbook/simple_tess/plot.py
Executable file
@ -0,0 +1,29 @@
|
|||||||
|
"""
|
||||||
|
Plot the columns of the output files
|
||||||
|
"""
|
||||||
|
import sys
|
||||||
|
from matplotlib import pyplot as plt
|
||||||
|
from mpl_toolkits.basemap import Basemap
|
||||||
|
import numpy as np
|
||||||
|
|
||||||
|
# Set up a projection
|
||||||
|
bm = Basemap(projection='ortho', lon_0=0, lat_0=0,
|
||||||
|
resolution='l', area_thresh=10000)
|
||||||
|
|
||||||
|
# Load the data and make them into matrices
|
||||||
|
data = np.loadtxt(sys.argv[1], unpack=True)
|
||||||
|
shape = (int(sys.argv[2]), int(sys.argv[3]))
|
||||||
|
lon = data[0].reshape(shape)
|
||||||
|
lat = data[1].reshape(shape)
|
||||||
|
glon, glat = bm(lon, lat)
|
||||||
|
|
||||||
|
plt.figure(figsize=(14, 12))
|
||||||
|
for i, value in enumerate(data[3:]):
|
||||||
|
plt.subplot(3, 4, i + 1)
|
||||||
|
plt.title("Column %d" % (i + 4))
|
||||||
|
bm.drawcoastlines()
|
||||||
|
bm.drawmapboundary()
|
||||||
|
bm.contourf(glon, glat, value.reshape(shape), 15, cmap=plt.cm.RdBu_r)
|
||||||
|
plt.colorbar(orientation="horizontal", pad=0, aspect=30)
|
||||||
|
plt.tight_layout()
|
||||||
|
plt.savefig('output.png')
|
||||||
12
dep/tesseroids/cookbook/simple_tess/simple_tess.bat
Executable file
@ -0,0 +1,12 @@
|
|||||||
|
:: Generate a regular grid, pipe it to all the computation programs,
|
||||||
|
:: and write the result to output.txt
|
||||||
|
|
||||||
|
tessgrd -r-45/45/-45/45 -b101/101 -z260e03 | ^
|
||||||
|
tesspot model.txt | ^
|
||||||
|
tessgx model.txt | tessgy model.txt | tessgz model.txt | ^
|
||||||
|
tessgxx model.txt | tessgxy model.txt | ^
|
||||||
|
tessgxz model.txt | tessgyy model.txt | ^
|
||||||
|
tessgyz model.txt | tessgzz model.txt -v -llog.txt > output.txt
|
||||||
|
|
||||||
|
:: Make a plot with the columns of output.txt
|
||||||
|
python plot.py output.txt 101 101
|
||||||
BIN
dep/tesseroids/cookbook/simple_tess/simple_tess.png
Executable file
|
After Width: | Height: | Size: 389 KiB |
14
dep/tesseroids/cookbook/simple_tess/simple_tess.sh
Executable file
@ -0,0 +1,14 @@
|
|||||||
|
#!/bin/bash
|
||||||
|
|
||||||
|
# Generate a regular grid, pipe it to all the computation programs,
|
||||||
|
# and write the result to output.txt
|
||||||
|
|
||||||
|
tessgrd -r-45/45/-45/45 -b101/101 -z260e03 | \
|
||||||
|
tesspot model.txt | \
|
||||||
|
tessgx model.txt | tessgy model.txt | tessgz model.txt | \
|
||||||
|
tessgxx model.txt | tessgxy model.txt | \
|
||||||
|
tessgxz model.txt | tessgyy model.txt | \
|
||||||
|
tessgyz model.txt | tessgzz model.txt -v -llog.txt > output.txt
|
||||||
|
|
||||||
|
# Make a plot with the columns of output.txt
|
||||||
|
python plot.py output.txt 101 101
|
||||||
32
dep/tesseroids/cookbook/tess2prism/plot.py
Executable file
@ -0,0 +1,32 @@
|
|||||||
|
"""
|
||||||
|
Plot the columns of the output files
|
||||||
|
"""
|
||||||
|
import sys
|
||||||
|
import pylab
|
||||||
|
from mpl_toolkits.basemap import Basemap
|
||||||
|
|
||||||
|
# Set up a projection
|
||||||
|
bm = Basemap(projection='ortho', lon_0=-80, lat_0=-40,
|
||||||
|
resolution='l', area_thresh=10000)
|
||||||
|
|
||||||
|
data = pylab.loadtxt(sys.argv[1], unpack=True)
|
||||||
|
shape = (int(sys.argv[2]), int(sys.argv[3]))
|
||||||
|
lon = pylab.reshape(data[0], shape)
|
||||||
|
lat = pylab.reshape(data[1], shape)
|
||||||
|
glon, glat = bm(lon, lat)
|
||||||
|
|
||||||
|
for i, value in enumerate(data[3:]):
|
||||||
|
value = pylab.reshape(value, shape)
|
||||||
|
pylab.figure(figsize=(4, 3))
|
||||||
|
pylab.title("Column %d" % (i + 4))
|
||||||
|
bm.drawcoastlines()
|
||||||
|
#bm.fillcontinents(color='coral',lake_color='aqua')
|
||||||
|
#bm.drawmapboundary(fill_color='aqua')
|
||||||
|
bm.drawmapboundary()
|
||||||
|
bm.drawparallels(pylab.arange(-90.,120.,30.))
|
||||||
|
bm.drawmeridians(pylab.arange(0.,420.,60.))
|
||||||
|
#bm.bluemarble()
|
||||||
|
bm.pcolor(glon, glat, value)
|
||||||
|
pylab.colorbar()
|
||||||
|
#bm.contour(glon, glat, value, 12, linewidth=3)
|
||||||
|
pylab.savefig('column%d.png' % (i + 4))
|
||||||
134
dep/tesseroids/cookbook/tess2prism/result.svg
Executable file
@ -0,0 +1,134 @@
|
|||||||
|
<?xml version="1.0" encoding="UTF-8" standalone="no"?>
|
||||||
|
<!-- Created with Inkscape (http://www.inkscape.org/) -->
|
||||||
|
|
||||||
|
<svg
|
||||||
|
xmlns:dc="http://purl.org/dc/elements/1.1/"
|
||||||
|
xmlns:cc="http://creativecommons.org/ns#"
|
||||||
|
xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
|
||||||
|
xmlns:svg="http://www.w3.org/2000/svg"
|
||||||
|
xmlns="http://www.w3.org/2000/svg"
|
||||||
|
xmlns:xlink="http://www.w3.org/1999/xlink"
|
||||||
|
xmlns:sodipodi="http://sodipodi.sourceforge.net/DTD/sodipodi-0.dtd"
|
||||||
|
xmlns:inkscape="http://www.inkscape.org/namespaces/inkscape"
|
||||||
|
width="1088.5714"
|
||||||
|
height="1122.8572"
|
||||||
|
id="svg2"
|
||||||
|
version="1.1"
|
||||||
|
inkscape:version="0.48.2 r9819"
|
||||||
|
sodipodi:docname="result.svg"
|
||||||
|
inkscape:export-filename="/home/leo/src/tesseroids/dev/cookbook/tess2prism/tess2prism.png"
|
||||||
|
inkscape:export-xdpi="90"
|
||||||
|
inkscape:export-ydpi="90">
|
||||||
|
<defs
|
||||||
|
id="defs4" />
|
||||||
|
<sodipodi:namedview
|
||||||
|
id="base"
|
||||||
|
pagecolor="#ffffff"
|
||||||
|
bordercolor="#666666"
|
||||||
|
borderopacity="1.0"
|
||||||
|
inkscape:pageopacity="1"
|
||||||
|
inkscape:pageshadow="2"
|
||||||
|
inkscape:zoom="0.35"
|
||||||
|
inkscape:cx="182.14285"
|
||||||
|
inkscape:cy="492.85716"
|
||||||
|
inkscape:document-units="px"
|
||||||
|
inkscape:current-layer="layer1"
|
||||||
|
showgrid="false"
|
||||||
|
inkscape:window-width="1680"
|
||||||
|
inkscape:window-height="1003"
|
||||||
|
inkscape:window-x="0"
|
||||||
|
inkscape:window-y="25"
|
||||||
|
inkscape:window-maximized="1"
|
||||||
|
fit-margin-top="0"
|
||||||
|
fit-margin-left="0"
|
||||||
|
fit-margin-right="0"
|
||||||
|
fit-margin-bottom="0" />
|
||||||
|
<metadata
|
||||||
|
id="metadata7">
|
||||||
|
<rdf:RDF>
|
||||||
|
<cc:Work
|
||||||
|
rdf:about="">
|
||||||
|
<dc:format>image/svg+xml</dc:format>
|
||||||
|
<dc:type
|
||||||
|
rdf:resource="http://purl.org/dc/dcmitype/StillImage" />
|
||||||
|
<dc:title></dc:title>
|
||||||
|
</cc:Work>
|
||||||
|
</rdf:RDF>
|
||||||
|
</metadata>
|
||||||
|
<g
|
||||||
|
inkscape:label="Layer 1"
|
||||||
|
inkscape:groupmode="layer"
|
||||||
|
id="layer1"
|
||||||
|
transform="translate(165.71428,783.35211)">
|
||||||
|
<image
|
||||||
|
y="-509.06641"
|
||||||
|
x="522.85712"
|
||||||
|
id="image3026"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column7.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="-234.78065"
|
||||||
|
x="522.85712"
|
||||||
|
id="image3059"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column10.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="39.505058"
|
||||||
|
x="522.85712"
|
||||||
|
id="image3092"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column13.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="-783.35211"
|
||||||
|
x="179.99997"
|
||||||
|
id="image2993"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column4.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="-509.06641"
|
||||||
|
x="179.99997"
|
||||||
|
id="image3015"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column6.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="-234.78065"
|
||||||
|
x="179.99997"
|
||||||
|
id="image3048"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column9.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="39.505058"
|
||||||
|
x="179.99997"
|
||||||
|
id="image3081"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column12.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="-509.06641"
|
||||||
|
x="-165.71428"
|
||||||
|
id="image3004"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column5.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="-234.78065"
|
||||||
|
x="-165.71428"
|
||||||
|
id="image3037"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column8.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
<image
|
||||||
|
y="39.505058"
|
||||||
|
x="-165.71428"
|
||||||
|
id="image3070"
|
||||||
|
xlink:href="file:///home/leo/src/tesseroids/dev/cookbook/tess2prism/column11.png"
|
||||||
|
height="300"
|
||||||
|
width="400" />
|
||||||
|
</g>
|
||||||
|
</svg>
|
||||||
|
After Width: | Height: | Size: 3.9 KiB |
11
dep/tesseroids/cookbook/tess2prism/sample-prism-model.txt
Executable file
@ -0,0 +1,11 @@
|
|||||||
|
# Prisms converted from tesseroid model with tess2prism 1.1dev
|
||||||
|
# local time: Wed May 16 14:34:47 2012
|
||||||
|
# tesseroids file: stdin
|
||||||
|
# conversion type: equal mass|spherical coordinates
|
||||||
|
# format: dx dy dz density lon lat r
|
||||||
|
# Test tesseroid model file
|
||||||
|
221766.31696055 169882.854778591 50000 499.977196258595 -76 -40 6378137
|
||||||
|
221766.31696055 169882.854778591 50000 499.977196258595 -78 -40 6378137
|
||||||
|
221766.31696055 169882.854778591 50000 499.977196258595 -80 -40 6378137
|
||||||
|
221766.31696055 169882.854778591 50000 499.977196258595 -82 -40 6378137
|
||||||
|
221766.31696055 169882.854778591 50000 499.977196258595 -84 -40 6378137
|
||||||
6
dep/tesseroids/cookbook/tess2prism/tess-model.txt
Executable file
@ -0,0 +1,6 @@
|
|||||||
|
# Test tesseroid model file
|
||||||
|
-77 -75 -41 -39 0 -50000 500
|
||||||
|
-79 -77 -41 -39 0 -50000 500
|
||||||
|
-81 -79 -41 -39 0 -50000 500
|
||||||
|
-83 -81 -41 -39 0 -50000 500
|
||||||
|
-85 -83 -41 -39 0 -50000 500
|
||||||
21
dep/tesseroids/cookbook/tess2prism/tess2prism.bat
Executable file
@ -0,0 +1,21 @@
|
|||||||
|
|
||||||
|
|
||||||
|
:: Generate a prism model from a tesseroid model.
|
||||||
|
:: Prisms will have the same mass as the tesseroids and
|
||||||
|
:: associated spherical coordinates of the center of
|
||||||
|
:: the top of the tesseroid.
|
||||||
|
|
||||||
|
tess2prism.exe < tess-model.txt > prism-model.txt
|
||||||
|
|
||||||
|
:: Generate a regular grid in spherical coordinates,
|
||||||
|
:: pipe the grid to the computation programs,
|
||||||
|
:: and dump the result on output.txt
|
||||||
|
:: prismpots calculates the potential in spherical
|
||||||
|
:: coordinates, prismgs calculates the full
|
||||||
|
:: gravity vector, and prismggts calculates the full
|
||||||
|
:: gravity gradient tensor.
|
||||||
|
|
||||||
|
tessgrd -r-160/0/-80/0 -b100/100 -z250e03 | ^
|
||||||
|
prismpots prism-model.txt | ^
|
||||||
|
prismgs prism-model.txt | ^
|
||||||
|
prismggts prism-model.txt -v > output.txt
|
||||||
BIN
dep/tesseroids/cookbook/tess2prism/tess2prism.png
Executable file
|
After Width: | Height: | Size: 888 KiB |
21
dep/tesseroids/cookbook/tess2prism/tess2prism.sh
Executable file
@ -0,0 +1,21 @@
|
|||||||
|
#!/bin/bash
|
||||||
|
|
||||||
|
# Generate a prism model from a tesseroid model.
|
||||||
|
# Prisms will have the same mass as the tesseroids and
|
||||||
|
# associated spherical coordinates of the center of
|
||||||
|
# the top of the tesseroid.
|
||||||
|
|
||||||
|
tess2prism < tess-model.txt > prism-model.txt
|
||||||
|
|
||||||
|
# Generate a regular grid in spherical coordinates,
|
||||||
|
# pipe the grid to the computation programs,
|
||||||
|
# and dump the result on output.txt
|
||||||
|
# prismpots calculates the potential in spherical
|
||||||
|
# coordinates, prismgs calculates the full
|
||||||
|
# gravity vector, and prismggts calculates the full
|
||||||
|
# gravity gradient tensor.
|
||||||
|
|
||||||
|
tessgrd -r-160/0/-80/0 -b100/100 -z250e03 | \
|
||||||
|
prismpots prism-model.txt | \
|
||||||
|
prismgs prism-model.txt | \
|
||||||
|
prismggts prism-model.txt -v > output.txt
|
||||||
25
dep/tesseroids/cookbook/tess2prism_flatten/plot.py
Executable file
@ -0,0 +1,25 @@
|
|||||||
|
"""
|
||||||
|
Plot the columns of the output files
|
||||||
|
"""
|
||||||
|
import sys
|
||||||
|
import pylab
|
||||||
|
|
||||||
|
data = pylab.loadtxt(sys.argv[1], unpack=True)
|
||||||
|
shape = (int(sys.argv[2]), int(sys.argv[3]))
|
||||||
|
lon = pylab.reshape(data[0], shape)*0.001
|
||||||
|
lat = pylab.reshape(data[1], shape)*0.001
|
||||||
|
xmin, xmax = lon.min(), lon.max()
|
||||||
|
ymin, ymax = lat.min(), lat.max()
|
||||||
|
for i, value in enumerate(data[3:]):
|
||||||
|
value = pylab.reshape(value, shape)
|
||||||
|
pylab.figure(figsize=(4, 3))
|
||||||
|
pylab.title("Column %d" % (i + 4))
|
||||||
|
pylab.axis('scaled')
|
||||||
|
pylab.pcolor(lon, lat, value)
|
||||||
|
pylab.colorbar()
|
||||||
|
pylab.contour(lon, lat, value, 12, color='k')
|
||||||
|
#pylab.xlabel("Longitude")
|
||||||
|
#pylab.ylabel("Latitude")
|
||||||
|
pylab.xlim(xmin, xmax)
|
||||||
|
pylab.ylim(ymin, ymax)
|
||||||
|
pylab.savefig('column%d.png' % (i + 4))
|
||||||
9
dep/tesseroids/cookbook/tess2prism_flatten/sample-prism-model.txt
Executable file
@ -0,0 +1,9 @@
|
|||||||
|
# Prisms converted from tesseroid model with tess2prism 1.1dev
|
||||||
|
# local time: Tue May 8 14:55:02 2012
|
||||||
|
# tesseroids file: stdin
|
||||||
|
# conversion type: flatten
|
||||||
|
# format: x1 x2 y1 y2 z1 z2 density
|
||||||
|
# Test tesseroid model file
|
||||||
|
1111100 1666650 1111100 1666650 0 30000 487.534658568521
|
||||||
|
-1111100 1111100 -1666650 -1111100 0 50000 198.175508383774
|
||||||
|
-1777760 -1111100 -1666650 555550 0 30000 -291.9029748328
|
||||||
4
dep/tesseroids/cookbook/tess2prism_flatten/tess-model.txt
Executable file
@ -0,0 +1,4 @@
|
|||||||
|
# Test tesseroid model file
|
||||||
|
10 15 10 15 0 -30000 500
|
||||||
|
-15 -10 -10 10 0 -50000 200
|
||||||
|
-15 5 -16 -10 0 -30000 -300
|
||||||
21
dep/tesseroids/cookbook/tess2prism_flatten/tess2prism_flatten.bat
Executable file
@ -0,0 +1,21 @@
|
|||||||
|
|
||||||
|
|
||||||
|
:: Generate a prism model from a tesseroid model by
|
||||||
|
:: flattening the tesseroids (1 degree = 111.11 km).
|
||||||
|
:: This way the converted prisms can be used
|
||||||
|
:: with the prism* programs in Cartesian coordinates.
|
||||||
|
|
||||||
|
tess2prism --flatten < tess-model.txt > prism-model.txt
|
||||||
|
|
||||||
|
:: Generate a regular grid in Cartesian coordinates,
|
||||||
|
:: pipe the grid to the computation programs,
|
||||||
|
:: and dump the result on output.txt
|
||||||
|
|
||||||
|
tessgrd -r-3e06/3e06/-3e06/3e06 -b50/50 -z250e03 | ^
|
||||||
|
prismpot prism-model.txt | ^
|
||||||
|
prismgx prism-model.txt | ^
|
||||||
|
prismgy prism-model.txt | ^
|
||||||
|
prismgz prism-model.txt | ^
|
||||||
|
prismgxx prism-model.txt | prismgxy prism-model.txt | ^
|
||||||
|
prismgxz prism-model.txt | prismgyy prism-model.txt | ^
|
||||||
|
prismgyz prism-model.txt | prismgzz prism-model.txt > output.txt
|
||||||
BIN
dep/tesseroids/cookbook/tess2prism_flatten/tess2prism_flatten.png
Executable file
|
After Width: | Height: | Size: 575 KiB |
21
dep/tesseroids/cookbook/tess2prism_flatten/tess2prism_flatten.sh
Executable file
@ -0,0 +1,21 @@
|
|||||||
|
#!/bin/bash
|
||||||
|
|
||||||
|
# Generate a prism model from a tesseroid model by
|
||||||
|
# flattening the tesseroids (1 degree = 111.11 km).
|
||||||
|
# This way the converted prisms can be used
|
||||||
|
# with the prism* programs in Cartesian coordinates.
|
||||||
|
|
||||||
|
tess2prism --flatten < tess-model.txt > prism-model.txt
|
||||||
|
|
||||||
|
# Generate a regular grid in Cartesian coordinates,
|
||||||
|
# pipe the grid to the computation programs,
|
||||||
|
# and dump the result on output.txt
|
||||||
|
|
||||||
|
tessgrd -r-3e06/3e06/-3e06/3e06 -b50/50 -z250e03 | \
|
||||||
|
prismpot prism-model.txt | \
|
||||||
|
prismgx prism-model.txt | \
|
||||||
|
prismgy prism-model.txt | \
|
||||||
|
prismgz prism-model.txt | \
|
||||||
|
prismgxx prism-model.txt | prismgxy prism-model.txt | \
|
||||||
|
prismgxz prism-model.txt | prismgyy prism-model.txt | \
|
||||||
|
prismgyz prism-model.txt | prismgzz prism-model.txt > output.txt
|
||||||
BIN
dep/tesseroids/cookbook/tesslayers/layers.png
Executable file
|
After Width: | Height: | Size: 45 KiB |
1683
dep/tesseroids/cookbook/tesslayers/layers.txt
Executable file
34
dep/tesseroids/cookbook/tesslayers/makelayers.py
Executable file
@ -0,0 +1,34 @@
|
|||||||
|
import numpy as np
|
||||||
|
import fatiando as ft
|
||||||
|
|
||||||
|
shape = (41, 41)
|
||||||
|
x, y = ft.grd.regular((-10, 10, 30, 50), shape)
|
||||||
|
height = 800 - 1000*ft.utils.gaussian2d(x, y, 3, 1, x0=0, y0=37)
|
||||||
|
rel = -7000*ft.utils.gaussian2d(x, y, 3, 5, x0=0, y0=40)
|
||||||
|
thick = height - rel
|
||||||
|
dens = 1900*np.ones_like(thick)
|
||||||
|
data = np.transpose([x, y, height, thick, dens])
|
||||||
|
with open('layers.txt', 'w') as f:
|
||||||
|
f.write("# Synthetic layer model of sediments and topography\n")
|
||||||
|
f.write("# Columns are:\n")
|
||||||
|
f.write("# lon lat height thickness density\n")
|
||||||
|
np.savetxt(f, data, fmt='%g')
|
||||||
|
ft.vis.figure(figsize=(4, 3))
|
||||||
|
ft.vis.title('Depth of sediments [m]')
|
||||||
|
ft.vis.axis('scaled')
|
||||||
|
ft.vis.pcolor(x, y, rel, shape)
|
||||||
|
ft.vis.colorbar()
|
||||||
|
ft.vis.savefig('depth.png')
|
||||||
|
ft.vis.figure(figsize=(4, 3))
|
||||||
|
ft.vis.title('Topography [m]')
|
||||||
|
ft.vis.axis('scaled')
|
||||||
|
ft.vis.pcolor(x, y, height, shape)
|
||||||
|
ft.vis.colorbar()
|
||||||
|
ft.vis.savefig('topography.png')
|
||||||
|
ft.vis.figure(figsize=(4, 3))
|
||||||
|
ft.vis.title('Thickness of sediment layer [m]')
|
||||||
|
ft.vis.axis('scaled')
|
||||||
|
ft.vis.pcolor(x, y, thick, shape)
|
||||||
|
ft.vis.colorbar()
|
||||||
|
ft.vis.savefig('thickness.png')
|
||||||
|
ft.vis.show()
|
||||||
20
dep/tesseroids/cookbook/tesslayers/plot.py
Executable file
@ -0,0 +1,20 @@
|
|||||||
|
"""
|
||||||
|
Plot the columns of the output files
|
||||||
|
"""
|
||||||
|
import sys
|
||||||
|
import pylab
|
||||||
|
|
||||||
|
data = pylab.loadtxt(sys.argv[1], unpack=True)
|
||||||
|
shape = (int(sys.argv[2]), int(sys.argv[3]))
|
||||||
|
lon = pylab.reshape(data[0], shape)
|
||||||
|
lat = pylab.reshape(data[1], shape)
|
||||||
|
for i, value in enumerate(data[3:]):
|
||||||
|
value = pylab.reshape(value, shape)
|
||||||
|
pylab.figure(figsize=(4, 3))
|
||||||
|
pylab.axis('scaled')
|
||||||
|
pylab.title("Column %d" % (i + 4))
|
||||||
|
pylab.pcolor(lon, lat, value)
|
||||||
|
pylab.colorbar()
|
||||||
|
pylab.xlim(lon.min(), lon.max())
|
||||||
|
pylab.ylim(lat.min(), lat.max())
|
||||||
|
pylab.savefig('column%d.png' % (i + 4))
|
||||||